

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.4 - phase: 0
(374 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
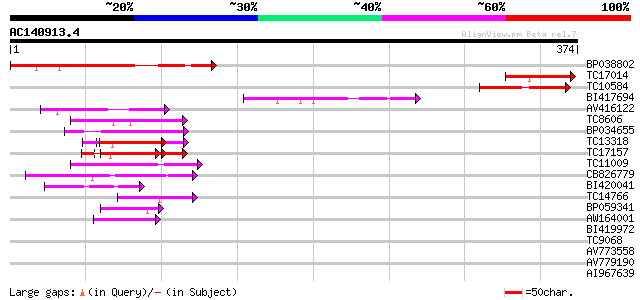
Score E
Sequences producing significant alignments: (bits) Value
BP038802 134 3e-32
TC17014 similar to UP|Q9SH58 (Q9SH58) F22C12.16, partial (11%) 77 4e-15
TC10584 similar to UP|Q9SH58 (Q9SH58) F22C12.16, partial (6%) 65 2e-11
BI417694 64 4e-11
AV416122 50 6e-07
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 49 1e-06
BP034655 49 2e-06
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 47 4e-06
TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly pr... 47 7e-06
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 46 9e-06
CB826779 41 3e-04
BI420041 41 4e-04
TC14766 similar to UP|Q7XXR8 (Q7XXR8) Nascent polypeptide associ... 41 4e-04
BP059341 40 6e-04
AW164001 40 6e-04
BI419972 39 0.001
TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 gluc... 39 0.001
AV773558 38 0.002
AV779190 38 0.003
AI967639 37 0.004
>BP038802
Length = 533
Score = 134 bits (336), Expect = 3e-32
Identities = 75/150 (50%), Positives = 94/150 (62%), Gaps = 14/150 (9%)
Frame = +1
Query: 1 MEAFSLLKYWRGGGAV---VGLSPSSDSTTTTNV-----------NTTTILTTAESSDDN 46
MEAFSLLKYWRGGG + L+PSS S TTT + NTT +T++ DD
Sbjct: 133 MEAFSLLKYWRGGGGAGPGLLLTPSSSSDTTTTISTTAAAVAQTDNTTDSSSTSDDDDDG 312
Query: 47 DDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKI 106
D+GPFFDLEFT P+E EDG + NH + ++EEE DE GE E EFK
Sbjct: 313 GDDGPFFDLEFTVPDESEDGQQNPNHTHPEEEEECDE--------------GEGEGEFKF 450
Query: 107 TLSPSNNERSDANLSLSPSEDLFFKGKLVQ 136
TLSPS+N+R+ S+SPS+DLFFKGK+V+
Sbjct: 451 TLSPSSNDRA---ASVSPSDDLFFKGKIVE 531
>TC17014 similar to UP|Q9SH58 (Q9SH58) F22C12.16, partial (11%)
Length = 719
Score = 77.4 bits (189), Expect = 4e-15
Identities = 41/48 (85%), Positives = 42/48 (87%), Gaps = 2/48 (4%)
Frame = +3
Query: 328 SASSAVAASPPVLV--SSKRRDDSLLQQQDGIQGAILHCKRSFNASRG 373
SASSAVAASPP SSKRRDDSLLQQQD IQGAILHC+RSFNASRG
Sbjct: 3 SASSAVAASPPAAAAESSKRRDDSLLQQQDAIQGAILHCQRSFNASRG 146
>TC10584 similar to UP|Q9SH58 (Q9SH58) F22C12.16, partial (6%)
Length = 480
Score = 65.1 bits (157), Expect = 2e-11
Identities = 32/60 (53%), Positives = 41/60 (68%)
Frame = +1
Query: 311 NIPAGLRVVYKHLGKSRSASSAVAASPPVLVSSKRRDDSLLQQQDGIQGAILHCKRSFNA 370
+ P G+RVV KHLGK RSA + V + P + R +DS+ Q DGIQ AILHC+RSFN+
Sbjct: 7 SFPVGMRVVSKHLGKRRSAGTVVGVNSP----ANRSNDSVQQHDDGIQSAILHCQRSFNS 174
>BI417694
Length = 436
Score = 63.9 bits (154), Expect = 4e-11
Identities = 45/130 (34%), Positives = 68/130 (51%), Gaps = 13/130 (10%)
Frame = +3
Query: 155 EPNNKPQFTASLLKSATKFRV--FMSGLKKPKTDSVLE------KKQHKPERI-----KF 201
EP +KPQ S+L+SA FR+ F G + KT+ + K Q K R+
Sbjct: 18 EPISKPQSPISILRSAPSFRISMFRRGKRMQKTEEPTKTECETMKDQKKETRVFAVKLNM 197
Query: 202 KVEEVPIVSLFTRDNSSKNSNKSQTNQNQKVEHESSLSPLEEKRFSKEIVMHKYLKMVKP 261
+E+ +RDNS+++ KV S + +RFSKE V+ KYLK++KP
Sbjct: 198 NMEDFNYSPALSRDNSTRSFGS-------KVRSFQSSEEQKPERFSKE-VLQKYLKLIKP 353
Query: 262 LYIRVSRRYS 271
LY++VS+RY+
Sbjct: 354 LYVKVSKRYT 383
>AV416122
Length = 414
Score = 50.1 bits (118), Expect = 6e-07
Identities = 30/89 (33%), Positives = 43/89 (47%), Gaps = 4/89 (4%)
Frame = +3
Query: 21 PSSDSTTTTN----VNTTTILTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQ 76
P+ DS N V+ T ++ NDD P D++F EEE+D
Sbjct: 174 PNPDSNDNNNNHPAVSATDDCSSQPKEAANDDNEPDNDVQFEGEEEEDD----------- 320
Query: 77 QEEEKDEDDDEDSDDDDDDDSGETESEFK 105
EE E++DE+ DD+DDD +GE E + K
Sbjct: 321 --EEDKEEEDEEEDDEDDDSNGEAEVDRK 401
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 48.9 bits (115), Expect = 1e-06
Identities = 28/102 (27%), Positives = 48/102 (46%), Gaps = 25/102 (24%)
Frame = -2
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGF-----------------------EETNHKNQQQ 77
E +D+D +GPF + E E+ G+ E N ++++
Sbjct: 596 EGDEDDDGDGPFGEGEEDLSSEDGAGYGNNSNNKSNSKKAPEGGPGAGAGPEENGEDEED 417
Query: 78 E--EEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSD 117
E E++++DDD+D DDD++DD GE E E + + +E D
Sbjct: 416 EDGEDQEDDDDDDEDDDEEDDGGEDEEEEGVEEEDNEDEEED 291
Score = 36.2 bits (82), Expect = 0.009
Identities = 15/51 (29%), Positives = 30/51 (58%)
Frame = -2
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDD 91
+ DD+DDE +EE+DG E+ + ++E+ +DE++DE+ ++
Sbjct: 401 QEDDDDDDEDD---------DEEDDGGEDEEEEGVEEEDNEDEEEDEEDEE 276
Score = 36.2 bits (82), Expect = 0.009
Identities = 13/54 (24%), Positives = 37/54 (68%)
Frame = -2
Query: 44 DDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDS 97
D+ D++G D E ++E+D E+ +++++E ++ED++++ +D++D+++
Sbjct: 428 DEEDEDGE--DQEDDDDDDEDDDEEDDGGEDEEEEGVEEEDNEDEEEDEEDEEA 273
Score = 35.0 bits (79), Expect = 0.020
Identities = 16/56 (28%), Positives = 31/56 (54%), Gaps = 12/56 (21%)
Frame = -2
Query: 60 PEEEEDGFEETNHKNQQQEEEKDEDDDEDSD------------DDDDDDSGETESE 103
PEE + E+ + ++Q+ +++ DEDDDE+ D +D++D+ + E E
Sbjct: 446 PEENGEDEEDEDGEDQEDDDDDDEDDDEEDDGGEDEEEEGVEEEDNEDEEEDEEDE 279
Score = 33.9 bits (76), Expect = 0.045
Identities = 13/43 (30%), Positives = 29/43 (67%)
Frame = -2
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDE 83
+ DD+DD+ + + +EEE+G EE ++++++++EE +E
Sbjct: 404 DQEDDDDDDEDDDEEDDGGEDEEEEGVEEEDNEDEEEDEEDEE 276
Score = 30.8 bits (68), Expect = 0.38
Identities = 17/58 (29%), Positives = 24/58 (41%), Gaps = 2/58 (3%)
Frame = -2
Query: 69 ETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKIT--LSPSNNERSDANLSLSP 124
++ K E E DD+ D DDD D GE E + NN + +N +P
Sbjct: 647 QSEDKQHDAEGEDGNDDEGDEDDDGDGPFGEGEEDLSSEDGAGYGNNSNNKSNSKKAP 474
>BP034655
Length = 517
Score = 48.5 bits (114), Expect = 2e-06
Identities = 25/82 (30%), Positives = 45/82 (54%)
Frame = +1
Query: 37 LTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDD 96
LT A+ +DDE EEEE+ EE + +++EEE++E+D+ED ++D+D+D
Sbjct: 109 LTAADFGLSDDDE-----------EEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDEDED 255
Query: 97 SGETESEFKITLSPSNNERSDA 118
+++ + N SD+
Sbjct: 256 YIPVAPSHRVSPNVHQNPTSDS 321
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 47.4 bits (111), Expect = 4e-06
Identities = 20/61 (32%), Positives = 36/61 (58%)
Frame = +3
Query: 58 TAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSD 117
T EEEED EE + +++ +++ D+DDD + ++DDDDD E E ++ + + +
Sbjct: 243 TDNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEVEVEGKDGAKGA 422
Query: 118 A 118
A
Sbjct: 423 A 425
Score = 47.0 bits (110), Expect = 5e-06
Identities = 21/55 (38%), Positives = 32/55 (58%)
Frame = +3
Query: 49 EGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
+GP ++ EEE+D EE + +++ D+DD E+ +DDDDDD E SE
Sbjct: 219 DGPPKPVQTDNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSE 383
Score = 46.6 bits (109), Expect = 7e-06
Identities = 20/48 (41%), Positives = 30/48 (61%), Gaps = 4/48 (8%)
Frame = +3
Query: 60 PEEEEDG----FEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
PE DG + N + + +EE+D++D++D DDDDDDD GE E +
Sbjct: 204 PEIYPDGPPKPVQTDNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEED 347
Score = 30.4 bits (67), Expect = 0.50
Identities = 16/65 (24%), Positives = 33/65 (50%)
Frame = +3
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGET 100
E DD+++E + + ++++DG +E++D+D DD++++ S E
Sbjct: 255 EEEDDDEEEDDEDEDDDDDDDDDDDG---------------EEEEDDDDDDEEEEGSEEV 389
Query: 101 ESEFK 105
E E K
Sbjct: 390 EVEGK 404
>TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly protein
1-like protein 3, partial (81%)
Length = 1298
Score = 46.6 bits (109), Expect = 7e-06
Identities = 17/43 (39%), Positives = 29/43 (66%)
Frame = +2
Query: 61 EEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
E E+ FE+ ++ +E++DEDDD+D +++DDDD E + E
Sbjct: 743 EAEQSDFEDIEEDDEDGDEDEDEDDDDDEEEEDDDDDDEDDEE 871
Score = 43.5 bits (101), Expect = 6e-05
Identities = 19/45 (42%), Positives = 27/45 (59%), Gaps = 2/45 (4%)
Frame = +2
Query: 57 FTAPEEEED--GFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGE 99
FT E+ D EE + + E+E D+DD+E+ DDDDDD+ E
Sbjct: 734 FTGEAEQSDFEDIEEDDEDGDEDEDEDDDDDEEEEDDDDDDEDDE 868
Score = 42.7 bits (99), Expect = 1e-04
Identities = 18/70 (25%), Positives = 45/70 (63%)
Frame = +2
Query: 48 DEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKIT 107
++ F D+E E++EDG E+ +++ +++++E+DD+D D+DD++ G+++S+
Sbjct: 749 EQSDFEDIE----EDDEDGDED---EDEDDDDDEEEEDDDDDDEDDEEGEGKSKSKSGSK 907
Query: 108 LSPSNNERSD 117
P ++ ++
Sbjct: 908 ARPGKDQPTE 937
Score = 32.3 bits (72), Expect = 0.13
Identities = 12/34 (35%), Positives = 20/34 (58%)
Frame = +2
Query: 70 TNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
T Q E+ +EDD++ +D+D+DD + E E
Sbjct: 737 TGEAEQSDFEDIEEDDEDGDEDEDEDDDDDEEEE 838
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 46.2 bits (108), Expect = 9e-06
Identities = 24/87 (27%), Positives = 45/87 (51%)
Frame = +2
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGET 100
+ DD+D G D + EEEE G E +++ D+DDD+D ++++++D G
Sbjct: 65 DEDDDDDAPGGGDDDDDEEDEEEEGGVEGGRGGGGDPDDDDDDDDDDDEEEEEEEDLG-- 238
Query: 101 ESEFKITLSPSNNERSDANLSLSPSED 127
+E+ + + + E +A+ P ED
Sbjct: 239 -TEYLVRRTVAAAEDEEASSDFEPEED 316
Score = 37.4 bits (85), Expect = 0.004
Identities = 31/89 (34%), Positives = 41/89 (45%), Gaps = 5/89 (5%)
Frame = +2
Query: 44 DDNDDEGPFFDLEFTAPEEEED-GFE----ETNHKNQQQEEEKDEDDDEDSDDDDDDDSG 98
DD+DD+ D E EEEED G E T + +E D + +ED DD+D+D G
Sbjct: 176 DDDDDDDDDDDEE---EEEEEDLGTEYLVRRTVAAAEDEEASSDFEPEEDEGDDNDNDDG 346
Query: 99 ETESEFKITLSPSNNERSDANLSLSPSED 127
E PS +RSD + S D
Sbjct: 347 EKAG------VPSKRKRSDKDGSGDDDSD 415
Score = 33.9 bits (76), Expect = 0.045
Identities = 15/58 (25%), Positives = 29/58 (49%)
Frame = +2
Query: 39 TAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDD 96
T +++D + F E + + D E+ ++++ +KD D+DSDD +DD
Sbjct: 260 TVAAAEDEEASSDFEPEEDEGDDNDNDDGEKAGVPSKRKRSDKDGSGDDDSDDGGEDD 433
>CB826779
Length = 469
Score = 41.2 bits (95), Expect = 3e-04
Identities = 33/117 (28%), Positives = 54/117 (45%), Gaps = 3/117 (2%)
Frame = +1
Query: 11 RGGGAVVGLSPSSDSTTT-TNVNTTTILTTAESSDDNDDEGPFF--DLEFTAPEEEEDGF 67
RG + L PS + + N + T ++S D+ + P D + P+E +D
Sbjct: 19 RGACLIAVLVPSPEEPHSHNNSQAMSPGTVFDASSDDHQQLPSATDDKKKPQPQEYDDA- 195
Query: 68 EETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSDANLSLSP 124
++ E ++DEDDDED +DD+D G TE K + S + ++ L L P
Sbjct: 196 --PVVEDVHDEADEDEDDDEDDEDDEDGAQGTTEGS-KQSRSEKKSRKAMLKLGLKP 357
>BI420041
Length = 452
Score = 40.8 bits (94), Expect = 4e-04
Identities = 25/66 (37%), Positives = 38/66 (56%)
Frame = +2
Query: 24 DSTTTTNVNTTTILTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDE 83
+S+ + V TI+ ++ DDE FD E +E+EDG E + + +QEEE+ E
Sbjct: 191 NSSDSEPVLEPTIVQEVSMDEEEDDE---FDYEEPLNDEDEDGDEY--YDDDEQEEEEFE 355
Query: 84 DDDEDS 89
D+DEDS
Sbjct: 356 DEDEDS 373
Score = 34.3 bits (77), Expect = 0.035
Identities = 27/87 (31%), Positives = 40/87 (45%), Gaps = 3/87 (3%)
Frame = +2
Query: 42 SSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSD---DDDDDDSG 98
SSD P E + EEE+D F+ EE D+DED D DDD+ +
Sbjct: 194 SSDSEPVLEPTIVQEVSMDEEEDDEFD---------YEEPLNDEDEDGDEYYDDDEQEEE 346
Query: 99 ETESEFKITLSPSNNERSDANLSLSPS 125
E E E + ++ + + S +SL+ S
Sbjct: 347 EFEDEDEDSVPYAGDGGSGGGISLAGS 427
>TC14766 similar to UP|Q7XXR8 (Q7XXR8) Nascent polypeptide associated
complex alpha chain, partial (58%)
Length = 600
Score = 40.8 bits (94), Expect = 4e-04
Identities = 19/58 (32%), Positives = 31/58 (52%), Gaps = 5/58 (8%)
Frame = +2
Query: 72 HKNQQQEEEKDEDDDEDSDDDDDDDS-----GETESEFKITLSPSNNERSDANLSLSP 124
H ++ E+ DEDDD++ DDD+DDD+ G+ K T S + ++ L + P
Sbjct: 116 HDDEPVVEDDDEDDDDEDDDDEDDDNIEGQEGDASGRSKQTRSEKKSRKAMLKLGMKP 289
>BP059341
Length = 471
Score = 40.0 bits (92), Expect = 6e-04
Identities = 20/52 (38%), Positives = 29/52 (55%), Gaps = 11/52 (21%)
Frame = -3
Query: 61 EEEEDGFEETNHKNQQQEEEKDEDDDEDS-----------DDDDDDDSGETE 101
EEEE+ + +N ++EEE D+DDD+ + DD DDDD GE +
Sbjct: 439 EEEEESSDFEPEENGEEEEEGDDDDDDKAEVPPKRKRSSKDDSDDDDGGEDD 284
Score = 32.0 bits (71), Expect = 0.17
Identities = 14/52 (26%), Positives = 25/52 (47%)
Frame = -3
Query: 45 DNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDD 96
+ ++E F+ E EEEE ++ + + ++ DD D DD +DD
Sbjct: 439 EEEEESSDFEPEENGEEEEEGDDDDDDKAEVPPKRKRSSKDDSDDDDGGEDD 284
Score = 29.6 bits (65), Expect = 0.85
Identities = 11/30 (36%), Positives = 19/30 (62%)
Frame = -3
Query: 66 GFEETNHKNQQQEEEKDEDDDEDSDDDDDD 95
G E ++ E E++ +++E+ DDDDDD
Sbjct: 448 GXAEEEEESSDFEPEENGEEEEEGDDDDDD 359
Score = 28.1 bits (61), Expect = 2.5
Identities = 14/38 (36%), Positives = 20/38 (51%)
Frame = -3
Query: 68 EETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFK 105
EE + + EE E+++E DDDDDD E + K
Sbjct: 439 EEEEESSDFEPEENGEEEEEG--DDDDDDKAEVPPKRK 332
>AW164001
Length = 447
Score = 40.0 bits (92), Expect = 6e-04
Identities = 18/44 (40%), Positives = 24/44 (53%)
Frame = +1
Query: 56 EFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGE 99
EF + D + N +EE+ DED D+D DDDDDDD +
Sbjct: 1 EFGTSDVVGDSVNKQNDDEVYKEEDVDEDFDDDDDDDDDDDDDD 132
Score = 38.5 bits (88), Expect = 0.002
Identities = 18/37 (48%), Positives = 25/37 (66%), Gaps = 2/37 (5%)
Frame = +1
Query: 74 NQQQEEE--KDEDDDEDSDDDDDDDSGETESEFKITL 108
N+Q ++E K+ED DED DDDDDDD + + + TL
Sbjct: 37 NKQNDDEVYKEEDVDEDFDDDDDDDDDDDDDDEPATL 147
Score = 37.4 bits (85), Expect = 0.004
Identities = 13/25 (52%), Positives = 20/25 (80%)
Frame = +1
Query: 72 HKNQQQEEEKDEDDDEDSDDDDDDD 96
+K + +E+ D+DDD+D DDDDDD+
Sbjct: 61 YKEEDVDEDFDDDDDDDDDDDDDDE 135
>BI419972
Length = 498
Score = 39.3 bits (90), Expect = 0.001
Identities = 15/31 (48%), Positives = 23/31 (73%)
Frame = +1
Query: 72 HKNQQQEEEKDEDDDEDSDDDDDDDSGETES 102
+K+ + E+++D DD++D DDDDDD GE S
Sbjct: 337 NKDSETEDDEDGDDEDDDADDDDDDEGEDYS 429
Score = 34.7 bits (78), Expect = 0.026
Identities = 11/26 (42%), Positives = 21/26 (80%)
Frame = +1
Query: 71 NHKNQQQEEEKDEDDDEDSDDDDDDD 96
N ++ +++E +D+D+D+DDDDDD+
Sbjct: 337 NKDSETEDDEDGDDEDDDADDDDDDE 414
Score = 31.2 bits (69), Expect = 0.29
Identities = 11/27 (40%), Positives = 18/27 (65%)
Frame = +1
Query: 77 QEEEKDEDDDEDSDDDDDDDSGETESE 103
++ E ++D+D D +DDD DD + E E
Sbjct: 340 KDSETEDDEDGDDEDDDADDDDDDEGE 420
Score = 30.4 bits (67), Expect = 0.50
Identities = 13/43 (30%), Positives = 25/43 (57%)
Frame = +1
Query: 61 EEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
E++EDG +E +++ D+DDD++ +D D+ E + E
Sbjct: 355 EDDEDGDDE--------DDDADDDDDDEGEDYSGDEGEEADPE 459
Score = 30.4 bits (67), Expect = 0.50
Identities = 10/36 (27%), Positives = 24/36 (65%)
Frame = +1
Query: 68 EETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
+++ ++ + +++D+D D+D DD+ +D SG+ E
Sbjct: 340 KDSETEDDEDGDDEDDDADDDDDDEGEDYSGDEGEE 447
Score = 26.9 bits (58), Expect = 5.5
Identities = 12/39 (30%), Positives = 22/39 (55%)
Frame = +1
Query: 79 EEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSD 117
++ + +DDED D D+DDD+ + + + S E +D
Sbjct: 340 KDSETEDDEDGD-DEDDDADDDDDDEGEDYSGDEGEEAD 453
>TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 glucosidase
SCW10 precursor (Soluble cell wall protein 10) ,
partial (6%)
Length = 1209
Score = 39.3 bits (90), Expect = 0.001
Identities = 17/46 (36%), Positives = 26/46 (55%)
Frame = +2
Query: 58 TAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
TAP GF+ +++ D+DDD+D DDDDDDD + + +
Sbjct: 41 TAPS----GFKRLFSPKPDDDDDDDDDDDDDDDDDDDDDDDDGDKK 166
Score = 38.1 bits (87), Expect = 0.002
Identities = 13/28 (46%), Positives = 21/28 (74%)
Frame = +2
Query: 78 EEEKDEDDDEDSDDDDDDDSGETESEFK 105
+++ D+DDD+D DDDDDDD + + + K
Sbjct: 83 DDDDDDDDDDDDDDDDDDDDDDDDGDKK 166
Score = 37.0 bits (84), Expect = 0.005
Identities = 12/30 (40%), Positives = 21/30 (70%)
Frame = +2
Query: 74 NQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
+ +++ D+DDD+D DDDDDDD + + +
Sbjct: 86 DDDDDDDDDDDDDDDDDDDDDDDGDKKQPD 175
Score = 36.2 bits (82), Expect = 0.009
Identities = 12/30 (40%), Positives = 21/30 (70%)
Frame = +2
Query: 74 NQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
+ +++ D+DDD+D DDDDD D + +S+
Sbjct: 92 DDDDDDDDDDDDDDDDDDDDDGDKKQPDSD 181
Score = 30.4 bits (67), Expect = 0.50
Identities = 11/43 (25%), Positives = 25/43 (57%)
Frame = +2
Query: 60 PEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETES 102
P++++D + +++ D+DDD+D DDD D ++++
Sbjct: 80 PDDDDD--------DDDDDDDDDDDDDDDDDDDGDKKQPDSDT 184
>AV773558
Length = 469
Score = 38.1 bits (87), Expect = 0.002
Identities = 26/67 (38%), Positives = 32/67 (46%)
Frame = -2
Query: 38 TTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDS 97
TT E S D G FF+ EE +D F E++ D+DDD+D DDD DD
Sbjct: 450 TTTEDSPD----GLFFN---EGDEELDDDFVLV-----YSEDDDDDDDDDDDDDDYTDDE 307
Query: 98 GETESEF 104
E E F
Sbjct: 306 DEEEGGF 286
Score = 26.2 bits (56), Expect = 9.4
Identities = 12/31 (38%), Positives = 20/31 (63%)
Frame = -2
Query: 36 ILTTAESSDDNDDEGPFFDLEFTAPEEEEDG 66
+L +E DD+DD+ D ++T E+EE+G
Sbjct: 381 VLVYSEDDDDDDDDDDDDD-DYTDDEDEEEG 292
>AV779190
Length = 539
Score = 37.7 bits (86), Expect = 0.003
Identities = 26/79 (32%), Positives = 36/79 (44%), Gaps = 19/79 (24%)
Frame = +3
Query: 42 SSDDNDDEGPFFDLEFTAPEEEE------DGFEETNHKNQQQEEEKDEDDDEDSDDDD-- 93
SS D+DD+ F++ E EEE D + + + E+E DDE+ DDDD
Sbjct: 228 SSVDSDDQSDFYEYEEERAEEESRKNKRYDPVFVNDELDSEIEDEIVPSDDENEDDDDDY 407
Query: 94 -----------DDDSGETE 101
DDDSGE +
Sbjct: 408 FGTKRNANAQSDDDSGEED 464
>AI967639
Length = 335
Score = 37.4 bits (85), Expect = 0.004
Identities = 19/69 (27%), Positives = 36/69 (51%)
Frame = +1
Query: 54 DLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNN 113
D+ ++ +EED E+ + ++EE+D+DDD+D D +DS +++ K+ P
Sbjct: 79 DISSSSEGDEEDEEEDQGDEYNLEDEEEDDDDDDDDDLSISEDS-DSDKPRKVKQLPGRT 255
Query: 114 ERSDANLSL 122
R S+
Sbjct: 256 RRETKRRSV 282
Score = 36.2 bits (82), Expect = 0.009
Identities = 17/55 (30%), Positives = 29/55 (51%), Gaps = 6/55 (10%)
Frame = +1
Query: 53 FDLEFTAPEEEEDGFEETNHKNQQQEEEKD------EDDDEDSDDDDDDDSGETE 101
+D E+ ++ ++ +++ EEE ED++ED DDDDDDD +E
Sbjct: 40 YDEEYLKKRKQRRDISSSSEGDEEDEEEDQGDEYNLEDEEEDDDDDDDDDLSISE 204
Score = 35.8 bits (81), Expect = 0.012
Identities = 17/55 (30%), Positives = 31/55 (55%)
Frame = +1
Query: 42 SSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDD 96
SS + D+E +EEED +E N ++++++++ D+DDD +D D D
Sbjct: 88 SSSEGDEE-----------DEEEDQGDEYNLEDEEEDDDDDDDDDLSISEDSDSD 219
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.304 0.123 0.325
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,252,703
Number of Sequences: 28460
Number of extensions: 68201
Number of successful extensions: 2207
Number of sequences better than 10.0: 217
Number of HSP's better than 10.0 without gapping: 855
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1371
length of query: 374
length of database: 4,897,600
effective HSP length: 92
effective length of query: 282
effective length of database: 2,279,280
effective search space: 642756960
effective search space used: 642756960
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC140913.4


