

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140849.12 + phase: 0
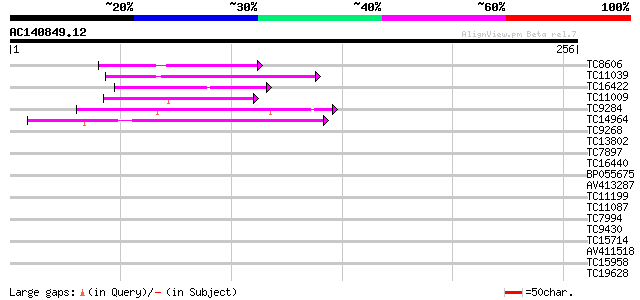
(256 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 45 1e-05
TC11039 weakly similar to UP|O65074 (O65074) YGL010w-like protei... 41 2e-04
TC16422 similar to PIR|T05653|T05653 amino acid transport protei... 40 4e-04
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 39 7e-04
TC9284 weakly similar to UP|Q9LPY0 (Q9LPY0) T23J18.22, partial (... 39 9e-04
TC14964 weakly similar to UP|SOX_ARATH (Q9SJA7) Potential sarcos... 39 9e-04
TC9268 similar to UP|Q9LHE6 (Q9LHE6) Genomic DNA, chromosome 3, ... 39 0.001
TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-... 39 0.001
TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-li... 39 0.001
TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinas... 37 0.003
BP055675 37 0.003
AV413287 37 0.003
TC11199 37 0.003
TC11087 weakly similar to UP|Q8GS92 (Q8GS92) OJ1513_F02.5 protei... 37 0.003
TC7994 similar to UP|Q8RU73 (Q8RU73) Ribose-5-phosphate isomeras... 37 0.003
TC9430 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase (L... 37 0.004
TC15714 weakly similar to UP|Q9LU96 (Q9LU96) Genomic DNA, chromo... 37 0.004
AV411518 37 0.004
TC15958 similar to GB|AAP37838.1|30725632|BT008479 At4g24220 {Ar... 37 0.004
TC19628 similar to PIR|T49142|T49142 CCR4-associated factor 1-li... 37 0.004
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 45.1 bits (105), Expect = 1e-05
Identities = 28/74 (37%), Positives = 45/74 (59%)
Frame = +1
Query: 41 GSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAW 100
G +SSS+S++ SSLS ++ +SS S+ PSS S + S+S S +S +++S+
Sbjct: 265 GCSASSSSSSSSSSSLSSSSTPSSSS----SSPPSSSSSSSSSSSSSSSWSSPSSSSSSS 432
Query: 101 GSSSRPSSASGPPT 114
SS P+ A GPP+
Sbjct: 433 PFSSGPAPAPGPPS 474
Score = 33.1 bits (74), Expect = 0.048
Identities = 26/72 (36%), Positives = 36/72 (49%)
Frame = +1
Query: 64 ASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSL 123
ASS S S+ S S T S+S S +S +++S SSS SS S P +S+ +S S
Sbjct: 274 ASSSSSSSSSSSLSSSSTPSSSSSSPPSSSSSSSS---SSSSSSSWSSPSSSSSSSPFSS 444
Query: 124 RPRSAETRPGSS 135
P A P +
Sbjct: 445 GPAPAPGPPSGA 480
Score = 32.7 bits (73), Expect = 0.063
Identities = 32/100 (32%), Positives = 48/100 (48%), Gaps = 4/100 (4%)
Frame = +1
Query: 32 PSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSD-- 89
PS + S SSSS+S++ SS S +++ +SSP P+ G PS + +
Sbjct: 328 PSSSSSSPPSSSSSSSSSSSSSSSWSSPSSSSSSSPFSSGPAPAPG----PPSGAFFELL 495
Query: 90 MASELTTTSAWGSSSRPSSAS--GPPTSNQTSQTSLRPRS 127
+ EL A S + SS S GP S+ +S +S P S
Sbjct: 496 LLFELLPYPAPSSDDKSSSPSPNGPSPSSSSSPSSSLPSS 615
Score = 32.0 bits (71), Expect = 0.11
Identities = 19/49 (38%), Positives = 31/49 (62%), Gaps = 1/49 (2%)
Frame = +1
Query: 91 ASELTTTSAWGSSSRPSS-ASGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
+S +++S+ SSS PSS +S PP+S+ +S +S S+ + P SS S
Sbjct: 283 SSSSSSSSSLSSSSTPSSSSSSPPSSSSSSSSSSSSSSSWSSPSSSSSS 429
Score = 29.6 bits (65), Expect = 0.53
Identities = 17/54 (31%), Positives = 30/54 (55%)
Frame = +1
Query: 77 GGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAET 130
G S + S+S S + ++T + SSS PSS+S +S+ +S + P S+ +
Sbjct: 265 GCSASSSSSSSSSSSLSSSSTPSSSSSSPPSSSSSSSSSSSSSSSWSSPSSSSS 426
Score = 26.2 bits (56), Expect = 5.9
Identities = 23/81 (28%), Positives = 31/81 (37%), Gaps = 1/81 (1%)
Frame = +2
Query: 64 ASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTS-QTS 122
A H S P+S T + GS T++S+WG++ P PP + Q
Sbjct: 158 AQEKKHGS*NPNS--YTTIQTGGGSTQLMLFTSSSSWGAAVLPHLPHPPPRLHYLPLQHP 331
Query: 123 LRPRSAETRPGSSHLSRFAEH 143
L P P HL R H
Sbjct: 332 LPPHLLHHHPLHRHLHRRHHH 394
>TC11039 weakly similar to UP|O65074 (O65074) YGL010w-like protein, partial
(79%)
Length = 733
Score = 40.8 bits (94), Expect = 2e-04
Identities = 29/97 (29%), Positives = 48/97 (48%)
Frame = +1
Query: 44 SSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSS 103
+S S S+++G S SP +++ S HLS+ PSS +++ S +S L +T W S
Sbjct: 97 TSQSTSSSFGQSSSPLSSSSTSH--HLSSLPSSPIIPLFSTSASSSQSSTLCSTPLWISK 270
Query: 104 SRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRF 140
PS S P ++ +S P ++ + L RF
Sbjct: 271 PVPSPRSSPFSAGSLQVSSQTPSASHSLGRLCWLPRF 381
>TC16422 similar to PIR|T05653|T05653 amino acid transport protein homolog
F22I13.20 - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (38%)
Length = 685
Score = 40.0 bits (92), Expect = 4e-04
Identities = 28/71 (39%), Positives = 35/71 (48%)
Frame = +3
Query: 48 ASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPS 107
+S+ +S P+ S PS SA PS+ S PST + L + SA SSSRP
Sbjct: 333 SSSPLAASSIPSLGTPNSDPSATSASPSAARSAASPSTP*LS-SPRLDSASATSSSSRPL 509
Query: 108 SASGPPTSNQT 118
SAS P T T
Sbjct: 510 SASSPATPPAT 542
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 39.3 bits (90), Expect = 7e-04
Identities = 27/71 (38%), Positives = 41/71 (57%), Gaps = 1/71 (1%)
Frame = -2
Query: 43 KSSSSASNAWGSSLSPNANAGASSPSHL-SARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
KSSSS+S++ SS S ++++G+ P S PSS S + S+S AS +++S
Sbjct: 234 KSSSSSSSSSSSSSSSSSSSGSPPPPRPPSTPPSSSSSSSSSSSSPPPGASSSSSSSPSS 55
Query: 102 SSSRPSSASGP 112
SS SS+S P
Sbjct: 54 SSPSSSSSSSP 22
Score = 37.4 bits (85), Expect = 0.003
Identities = 35/104 (33%), Positives = 47/104 (44%), Gaps = 5/104 (4%)
Frame = -2
Query: 37 TLGWGSKSSSSASNAWGSSLSPNA----NAGASSP-SHLSARPSSGGSGTRPSTSGSDMA 91
+L S SSSS S + +S S A S P S S+ SS S + S+SGS
Sbjct: 339 SLSLSSPSSSSGSKSLDASSSSAAATVRRTRYSVPKSSSSSSSSSSSSSSSSSSSGSPPP 160
Query: 92 SELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
+T SSS SS+S PP +S +S S+ + SS
Sbjct: 159 PRPPSTPPSSSSSSSSSSSSPPPGASSSSSSSPSSSSPSSSSSS 28
Score = 34.3 bits (77), Expect = 0.022
Identities = 26/90 (28%), Positives = 44/90 (48%), Gaps = 5/90 (5%)
Frame = -2
Query: 54 SSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASG-- 111
SSLS ++ + +S L A SS + R + +S +++S+ SSS SS+
Sbjct: 342 SSLSLSSPSSSSGSKSLDASSSSAAATVRRTRYSVPKSSSSSSSSSSSSSSSSSSSGSPP 163
Query: 112 ---PPTSNQTSQTSLRPRSAETRPGSSHLS 138
PP++ +S +S S+ PG+S S
Sbjct: 162 PPRPPSTPPSSSSSSSSSSSSPPPGASSSS 73
Score = 33.1 bits (74), Expect = 0.048
Identities = 34/106 (32%), Positives = 47/106 (44%), Gaps = 11/106 (10%)
Frame = -2
Query: 44 SSSSASNAWGSSLSPNAN-------AGASSPSHLSARPSSGGSGTRP-STSGSDMASELT 95
SSSS ++ SS P+ + A SPS LS S SG++ S S A+ +
Sbjct: 435 SSSSPPSSESSSPEPSLSDLFLFDGTPAFSPSSLSLSSPSSSSGSKSLDASSSSAAATVR 256
Query: 96 TTSAWGSSSRPSSASGPPTSNQTSQTS---LRPRSAETRPGSSHLS 138
T S SS+S +S+ +S +S PR T P SS S
Sbjct: 255 RTRYSVPKSSSSSSSSSSSSSSSSSSSGSPPPPRPPSTPPSSSSSS 118
Score = 27.7 bits (60), Expect = 2.0
Identities = 21/70 (30%), Positives = 36/70 (51%), Gaps = 7/70 (10%)
Frame = -2
Query: 30 NLPSQRGTLGWGSKSSSSASNAWGSS------LSPNANAGASSPSHLSARP-SSGGSGTR 82
++P + S SSSS+S++ GS +P +++ +SS S S P +S S +
Sbjct: 243 SVPKSSSSSSSSSSSSSSSSSSSGSPPPPRPPSTPPSSSSSSSSSSSSPPPGASSSSSSS 64
Query: 83 PSTSGSDMAS 92
PS+S +S
Sbjct: 63 PSSSSPSSSS 34
>TC9284 weakly similar to UP|Q9LPY0 (Q9LPY0) T23J18.22, partial (33%)
Length = 898
Score = 38.9 bits (89), Expect = 9e-04
Identities = 37/122 (30%), Positives = 53/122 (43%), Gaps = 4/122 (3%)
Frame = +1
Query: 31 LPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGAS-SPSHLSARPSSGGSGTRPSTSGSD 89
LP + G S S + + S N AS SPS+ P+S S T+PS + S
Sbjct: 70 LPLRSSHCGSSSPSYGAVCHRILLSTPTTTNESASFSPSNPDPNPASSASTTKPSAATSP 249
Query: 90 MASELTTTSAWGSSSRPSSASGPPTSN---QTSQTSLRPRSAETRPGSSHLSRFAEHVGE 146
+ T + SS+ SS + P +++ TS S P S+ T P H SR +
Sbjct: 250 PSKPHAPTPSSSSSTPLSSTTNPNSASPSFATSSPSSPPPSSPTTPTPPH-SRISSSATL 426
Query: 147 NS 148
NS
Sbjct: 427 NS 432
Score = 31.6 bits (70), Expect = 0.14
Identities = 25/68 (36%), Positives = 36/68 (52%), Gaps = 2/68 (2%)
Frame = +1
Query: 45 SSSASNAWGSSLSPNANAGASSPSHLSARPSS--GGSGTRPSTSGSDMASELTTTSAWGS 102
SSS+S S+ +PN ++SPS ++ PSS S T P+ S ++S T S S
Sbjct: 277 SSSSSTPLSSTTNPN----SASPSFATSSPSSPPPSSPTTPTPPHSRISSSATLNSRNRS 444
Query: 103 SSRPSSAS 110
P+SAS
Sbjct: 445 LVSPNSAS 468
>TC14964 weakly similar to UP|SOX_ARATH (Q9SJA7) Potential sarcosine oxidase
, partial (33%)
Length = 612
Score = 38.9 bits (89), Expect = 9e-04
Identities = 37/137 (27%), Positives = 56/137 (40%), Gaps = 1/137 (0%)
Frame = +1
Query: 9 TSSTRRGGMTVLGKVAVPKPINLP-SQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSP 67
T+S T G PK S+R TL W + S + LS + +SP
Sbjct: 205 TTSPSSSPPTTYGNKPRPKSATTSCSRRSTLTWPPPMTPSFT------LSSRIASATTSP 366
Query: 68 SHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRS 127
+ S SS S STS + S T+T+ S +PS S +N + PR
Sbjct: 367 MNSSTAASSPRSSPENSTSRTTGLSSPTSTAVSSSPPKPSPCSRRSPTNSAPCSRTTPR* 546
Query: 128 AETRPGSSHLSRFAEHV 144
+ +R + +SR ++ V
Sbjct: 547 STSRKNETAVSRSSQAV 597
>TC9268 similar to UP|Q9LHE6 (Q9LHE6) Genomic DNA, chromosome 3, P1 clone:
MZE19, partial (30%)
Length = 805
Score = 38.5 bits (88), Expect = 0.001
Identities = 29/81 (35%), Positives = 46/81 (55%), Gaps = 3/81 (3%)
Frame = +3
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGG-SGTRPSTSGSDMASELTTTSAW 100
+ S+S+AS+ G L+P+ +A +S+P L+ P+S S T + S A+ T SAW
Sbjct: 339 TSSTSTASSTSGKPLAPSPSAASSTP--LTPTPTSTSPSSTLQLPATSSAATSATKPSAW 512
Query: 101 --GSSSRPSSASGPPTSNQTS 119
S+S P+S S TS+ T+
Sbjct: 513 FTCSASCPASNSSTMTSSSTT 575
Score = 37.0 bits (84), Expect = 0.003
Identities = 32/105 (30%), Positives = 49/105 (46%), Gaps = 14/105 (13%)
Frame = +3
Query: 32 PSQRGTLGWGSK------SSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSG----T 81
PS R L W +S+ S A S+ S + A ++S L+ PS+ S T
Sbjct: 249 PSTRTYLRWSQSCNPPAPASAGTSTAASSNTSSTSTASSTSGKPLAPSPSAASSTPLTPT 428
Query: 82 RPSTSGSDMASELTTTSAWGSSSRPSS----ASGPPTSNQTSQTS 122
STS S T+SA S+++PS+ ++ P SN ++ TS
Sbjct: 429 PTSTSPSSTLQLPATSSAATSATKPSAWFTCSASCPASNSSTMTS 563
>TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-like
protein, partial (41%)
Length = 522
Score = 38.5 bits (88), Expect = 0.001
Identities = 36/107 (33%), Positives = 52/107 (47%)
Frame = +2
Query: 26 PKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPST 85
PKP P+ TL S S +SAS A + SP SSPS SA PSS + PST
Sbjct: 152 PKPSPSPAPSSTLRAPSSSPASASPASSPTKSPKP----SSPS-ASAPPSS--TPLTPST 310
Query: 86 SGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRP 132
+ S+ + T++S+ S++ PPTS +L ++ + P
Sbjct: 311 ATSESSPTATSSSS-------SASPAPPTSFSALSRALELKALASSP 430
Score = 31.6 bits (70), Expect = 0.14
Identities = 24/87 (27%), Positives = 41/87 (46%), Gaps = 5/87 (5%)
Frame = +2
Query: 54 SSLSPNAN---AGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPS--S 108
S +S N N + SSP+ + +PS + + + S + + S+ S +PS S
Sbjct: 89 SQISSNPNKTTSTTSSPTSTTPKPSPSPAPSSTLRAPSSSPASASPASSPTKSPKPSSPS 268
Query: 109 ASGPPTSNQTSQTSLRPRSAETRPGSS 135
AS PP+S + ++ S+ T SS
Sbjct: 269 ASAPPSSTPLTPSTATSESSPTATSSS 349
Score = 26.6 bits (57), Expect = 4.5
Identities = 25/61 (40%), Positives = 31/61 (49%), Gaps = 5/61 (8%)
Frame = +2
Query: 42 SKSSSSASNAWGSSLSP-----NANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTT 96
+ SSSSAS A +S S A ASSPS S+ P S S STS S +S +T
Sbjct: 338 TSSSSSASPAPPTSFSALSRALELKALASSPSPPSSPPPSPPSAICMSTSLSSASSVRST 517
Query: 97 T 97
+
Sbjct: 518 S 520
Score = 25.8 bits (55), Expect = 7.7
Identities = 17/74 (22%), Positives = 29/74 (38%)
Frame = +2
Query: 65 SSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLR 124
+ P H + + P+ + S +S +TT S PSS P+S+ S +
Sbjct: 53 NKPLHSTLTKPPSQISSNPNKTTSTTSSPTSTTPKPSPSPAPSSTLRAPSSSPASASPAS 232
Query: 125 PRSAETRPGSSHLS 138
+ +P S S
Sbjct: 233 SPTKSPKPSSPSAS 274
>TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-like protein
2 precursor (Phytocyanin-like protein), partial (31%)
Length = 1482
Score = 38.5 bits (88), Expect = 0.001
Identities = 38/119 (31%), Positives = 52/119 (42%), Gaps = 5/119 (4%)
Frame = +3
Query: 25 VPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANA---GASSPSH--LSARPSSGGS 79
+P P P G G+ S+S S + +P A + GA SPS S P+ G +
Sbjct: 645 LPSPAGAPVPAGGPSAGTPPSASTSPSPSPVSAPPAASPAPGAESPSSSPTSNSPAGGPT 824
Query: 80 GTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
T PS +G A S + P SASGP S + +SL A+T SS+ S
Sbjct: 825 ATSPSPAGDSPAGGPPAPSPT-TGDTPPSASGPDVSPSGTPSSL---PADTPSSSSNNS 989
Score = 33.9 bits (76), Expect = 0.028
Identities = 32/101 (31%), Positives = 43/101 (41%), Gaps = 10/101 (9%)
Frame = +3
Query: 26 PKPINLPSQRGTL-GWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSG------- 77
P P++ P G S SSS SN SP A+SPS P+ G
Sbjct: 726 PSPVSAPPAASPAPGAESPSSSPTSN------SPAGGPTATSPSPAGDSPAGGPPAPSPT 887
Query: 78 GSGTRPSTSGSDMASELTTTS--AWGSSSRPSSASGPPTSN 116
T PS SG D++ T +S A SS ++++ PP N
Sbjct: 888 TGDTPPSASGPDVSPSGTPSSLPADTPSSSSNNSTAPPPGN 1010
>TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinase, partial
(30%)
Length = 549
Score = 37.4 bits (85), Expect = 0.003
Identities = 39/127 (30%), Positives = 50/127 (38%), Gaps = 6/127 (4%)
Frame = +2
Query: 6 RRWTSSTRRGGMTVLGKVAVPKPINLPSQRGTLGW--GSKSSSSASNAWGSSLSPNANAG 63
RRWT RG L S + T W G +S+SS++ S SP + A
Sbjct: 197 RRWTPPWSRGSCEKLTPCDASSTTRT-SSKSTRSWPPGPRSTSSSTTPAEESSSPRSPAE 373
Query: 64 ASSPSHLSARPSSGGSGTRPSTSGSDMASELT----TTSAWGSSSRPSSASGPPTSNQTS 119
A+S S A SS S + PS S ++ AS TS+W P S
Sbjct: 374 AASRSPSPAATSS--SSSPPSVSATETASRTATSSRRTSSW-----------TPRETSRS 514
Query: 120 QTSLRPR 126
TS PR
Sbjct: 515 LTSASPR 535
Score = 27.3 bits (59), Expect = 2.6
Identities = 38/149 (25%), Positives = 56/149 (37%), Gaps = 18/149 (12%)
Frame = +2
Query: 8 WTSSTRRGGMTVLGKVAVPKPINLPSQRGTLGWGSKSSSSASN---AWG-SSLSPNANAG 63
W S++ RG G V + R W S+ S+S W S
Sbjct: 83 WGSTSSRGSW---GAVISQRCTKRCLSRTAPRWPSR*STSPRRWTPPWSRGSCEKLTPCD 253
Query: 64 ASSPSHLSARPSSGGSGTRPSTSGS--------------DMASELTTTSAWGSSSRPSSA 109
ASS + S++ + STS S + AS + +A SSS P S
Sbjct: 254 ASSTTRTSSKSTRSWPPGPRSTSSSTTPAEESSSPRSPAEAASRSPSPAATSSSSSPPSV 433
Query: 110 SGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
S T+++T+ +S R S R S L+
Sbjct: 434 SATETASRTATSSRRTSSWTPRETSRSLT 520
>BP055675
Length = 564
Score = 37.4 bits (85), Expect = 0.003
Identities = 46/175 (26%), Positives = 71/175 (40%), Gaps = 10/175 (5%)
Frame = +3
Query: 26 PKPINLPSQRGTLGWGSKSSSSASNAW-GSSLSPNANAGASSPSHLSARPSSGGSGTR-- 82
P P + P+ GT+G GS + + G+SL+P +A S P++ G GTR
Sbjct: 18 PAPSSTPASLGTIGIGSAPRNVNIHIHAGTSLAPIVSAIGSRPNN--------GEGTRSE 173
Query: 83 ----PSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQT---SQTSLRPRSAETRPGSS 135
P + S L +A S++ PS G + T S S ++ P SS
Sbjct: 174 HRNEPGSGDSGSTRVLPVRNA-VSATLPSHTPGVGIAGSTQTGSGVSTSQPPHDSAPFSS 350
Query: 136 HLSRFAEHVGENSVAWNGARTTETLGITQRKNDDFSLSTGDFPTLGSEKDKSVHD 190
L+ + NSV T G + N D S P LG+E+ ++ +
Sbjct: 351 VLAEIHSRL-RNSVGNMQGNNTVPSGQMESSNRDLSSGPESRPPLGNEQQDTMEN 512
>AV413287
Length = 255
Score = 37.0 bits (84), Expect = 0.003
Identities = 26/70 (37%), Positives = 36/70 (51%)
Frame = +1
Query: 41 GSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAW 100
G ++ SS++ A +S S ++ SSPSH S SS S STS S A T T W
Sbjct: 22 G*RAGSSSTTA--ASCSDQTHSTPSSPSHGSPSASSSPSSESTSTSLSLNALSATPTKCW 195
Query: 101 GSSSRPSSAS 110
SSS ++ +
Sbjct: 196 ASSSSSAATA 225
>TC11199
Length = 743
Score = 37.0 bits (84), Expect = 0.003
Identities = 31/82 (37%), Positives = 41/82 (49%), Gaps = 8/82 (9%)
Frame = +1
Query: 52 WGSSLSPNANAGASSPSHLSARPSSGGSGTR-PSTSGSDMA-----SELTTTSAWGSSSR 105
W SS SP A + SARPSS S + +TS + +A S+ + +SS
Sbjct: 109 WTSSSSPPPYATTRT*DPSSARPSSPASQSHSTTTSATSLAPKSPRSKRSARRTTRTSSS 288
Query: 106 PSSASGP--PTSNQTSQTSLRP 125
PS+ S P PTSN +S SL P
Sbjct: 289 PSTISDPSSPTSNPSSPPSLTP 354
Score = 32.3 bits (72), Expect = 0.082
Identities = 20/55 (36%), Positives = 28/55 (50%)
Frame = +1
Query: 85 TSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSR 139
TS S TT + SS+RPSS + S TS TSL P+S ++ + +R
Sbjct: 112 TSSSSPPPYATTRT*DPSSARPSSPASQSHSTTTSATSLAPKSPRSKRSARRTTR 276
>TC11087 weakly similar to UP|Q8GS92 (Q8GS92) OJ1513_F02.5 protein
(OJ1118_G09.13 protein), partial (6%)
Length = 689
Score = 37.0 bits (84), Expect = 0.003
Identities = 24/91 (26%), Positives = 42/91 (45%)
Frame = +1
Query: 44 SSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSS 103
S ++ A S+ SP+ + S S+ GS P+T+ ++ S +TS+ +
Sbjct: 28 SKTTTPMATWSTPSPSTPTPSKPSSPQIPPTSTPGSPKPPATATTNSPSSSVSTSSGAPT 207
Query: 104 SRPSSASGPPTSNQTSQTSLRPRSAETRPGS 134
+PS + P+SN S T+ + T P S
Sbjct: 208 PKPSRTTPSPSSNSASTTAASSSKSSTHPPS 300
>TC7994 similar to UP|Q8RU73 (Q8RU73) Ribose-5-phosphate isomerase
precursor , partial (81%)
Length = 1355
Score = 37.0 bits (84), Expect = 0.003
Identities = 34/110 (30%), Positives = 47/110 (41%), Gaps = 1/110 (0%)
Frame = +2
Query: 58 PNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSAS-GPPTSN 116
P N SPS + S TRPST+ S S + + SS PSSAS PP ++
Sbjct: 365 PLPNPSVPSPSPKTT--SRDSPPTRPSTTSSPAWSSASAPAPPPPSSSPSSASFSPPANS 538
Query: 117 QTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTETLGITQRK 166
TS S P + +R S + S++ + A T TL T +
Sbjct: 539 PTSSASPPPSARRSRRDPSASRSPSSTTIPASISPSTAPTRSTLSSTSSR 688
Score = 35.8 bits (81), Expect = 0.007
Identities = 34/118 (28%), Positives = 52/118 (43%)
Frame = +2
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
++ S+++S AW S+ +P +SSPS S P P+ S + AS SA
Sbjct: 428 TRPSTTSSPAWSSASAPAPPPPSSSPSSASFSP--------PANSPTSSAS--PPPSARR 577
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTET 159
S PS++ P S+ T S+ P +A TR S S + W+ R T +
Sbjct: 578 SRRDPSASRSP--SSTTIPASISPSTAPTRSTLSSTSSRVAAALFSGRRWSRPRLTNS 745
>TC9430 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase (LDH) ,
partial (49%)
Length = 866
Score = 36.6 bits (83), Expect = 0.004
Identities = 31/86 (36%), Positives = 40/86 (46%), Gaps = 4/86 (4%)
Frame = +1
Query: 54 SSLSPNANAGASSPSHLSARPSSGGS----GTRPSTSGSDMASELTTTSAWGSSSRPSSA 109
++LS A ASSP L+ P+S GS GT PS+S S W + R S+
Sbjct: 529 TTLSLPAPISASSPPELARSPASPGSTSCRGTSPSSSRS--------YPLWFVTPRTVSS 684
Query: 110 SGPPTSNQTSQTSLRPRSAETRPGSS 135
S PT + +S TSL GSS
Sbjct: 685 SSFPTPSMSSPTSLGSSPGSHPTGSS 762
>TC15714 weakly similar to UP|Q9LU96 (Q9LU96) Genomic DNA, chromosome 3, P1
clone: MPE11, partial (26%)
Length = 1157
Score = 36.6 bits (83), Expect = 0.004
Identities = 37/108 (34%), Positives = 51/108 (46%), Gaps = 10/108 (9%)
Frame = +2
Query: 42 SKSSSSASNAWGSSLSPNAN---AGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTS 98
S S+SSA++++ ++ +P + A ASSPS + PSS S S + +
Sbjct: 233 SPSTSSATSSFSATGAPRFDPNHAAASSPSSTAPPPSS---------SASPPSQTPVSPP 385
Query: 99 AWGSSSRPSSASGPPTSN---QTSQTSLRP----RSAETRPGSSHLSR 139
S PS S PPTS+ T+ SL P S T P SS LSR
Sbjct: 386 PTPRSRTPSLISAPPTSSPICYTTSLSLPPPTRSSSRTTSPRSSWLSR 529
>AV411518
Length = 360
Score = 36.6 bits (83), Expect = 0.004
Identities = 31/80 (38%), Positives = 41/80 (50%), Gaps = 3/80 (3%)
Frame = +3
Query: 45 SSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRP---STSGSDMASELTTTSAWG 101
+ SA +W S +P ++ G +SPS PSS RP STSG + T +
Sbjct: 126 AGSALASWASPCAPTSS-GPASPS-----PSSTAPSPRPNPSSTSGPTSPTPHTPSPPPP 287
Query: 102 SSSRPSSASGPPTSNQTSQT 121
+SS PSSA PPTS +S T
Sbjct: 288 TSSSPSSAI-PPTSAPSSST 344
>TC15958 similar to GB|AAP37838.1|30725632|BT008479 At4g24220 {Arabidopsis
thaliana;}, partial (52%)
Length = 800
Score = 36.6 bits (83), Expect = 0.004
Identities = 33/113 (29%), Positives = 53/113 (46%), Gaps = 16/113 (14%)
Frame = +3
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGS------------- 88
S +S +A + G+ ++ + + A+SP + +PSS S T P++S S
Sbjct: 339 STASPAALDPPGTPITQSTTSSATSPIPTTPKPSSPSSPTSPTSSTSPGPTATPNRRIAR 518
Query: 89 DMASELTTTSAWGSSSRPSSA---SGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
A T+SA S +RP SA S P S ++ +S RS+ T S +S
Sbjct: 519 STAPCSETSSAPSSPTRPISAT*RSKPAVSTTSALSSSSARSSSTSRRSRRIS 677
Score = 31.2 bits (69), Expect = 0.18
Identities = 30/90 (33%), Positives = 44/90 (48%)
Frame = +3
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S +S ++S + G + +PN A S + S S+ S TRP ++ ++TTSA
Sbjct: 450 SPTSPTSSTSPGPTATPNRRI-ARSTAPCSETSSAPSSPTRPISAT*RSKPAVSTTSALS 626
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETR 131
SSS SS+ TS+ S R A TR
Sbjct: 627 SSSARSSS--------TSRRSRRISPALTR 692
>TC19628 similar to PIR|T49142|T49142 CCR4-associated factor 1-like protein
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(30%)
Length = 537
Score = 36.6 bits (83), Expect = 0.004
Identities = 23/70 (32%), Positives = 35/70 (49%)
Frame = +2
Query: 55 SLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPT 114
+L P N +P S+ G + P+++GSD LT +S W SS P+S+S P
Sbjct: 152 NLYPLTNQPCPNPRDRSSPDQCGPPTSNPNSNGSDP*LMLTLSSPWTPSS-PASSSAPTP 328
Query: 115 SNQTSQTSLR 124
S T++R
Sbjct: 329 PTPPSTTAIR 358
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.125 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,060,854
Number of Sequences: 28460
Number of extensions: 79934
Number of successful extensions: 1254
Number of sequences better than 10.0: 301
Number of HSP's better than 10.0 without gapping: 1008
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1171
length of query: 256
length of database: 4,897,600
effective HSP length: 88
effective length of query: 168
effective length of database: 2,393,120
effective search space: 402044160
effective search space used: 402044160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC140849.12


