

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140722.2 + phase: 0
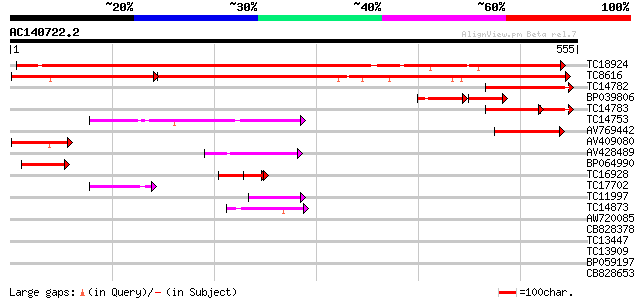
(555 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC18924 homologue to UP|Q94FN3 (Q94FN3) Phosphatidylinositol tra... 631 0.0
TC8616 UP|Q94FN1 (Q94FN1) Phosphatidylinositol transfer-like pro... 357 e-154
TC14782 UP|O04360 (O04360) Late nodulin Nlj16, complete 89 2e-18
BP039806 55 2e-17
TC14783 UP|O04360 (O04360) Late nodulin Nlj16, complete 62 7e-14
TC14753 similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial... 74 8e-14
AV769442 73 1e-13
AV409080 62 3e-10
AV428489 59 2e-09
BP064990 55 3e-08
TC16928 UP|Q94FN0 (Q94FN0) Phosphatidylinositol transfer-like pr... 49 2e-06
TC17702 weakly similar to GB|AAN73306.1|25141223|BT002309 At3g51... 49 3e-06
TC11997 similar to UP|O48940 (O48940) Polyphosphoinositide bindi... 45 3e-05
TC14873 weakly similar to GB|BAB08982.1|9758453|AB008271 seleniu... 44 5e-05
AW720085 38 0.004
CB828378 38 0.005
TC13447 37 0.006
TC13909 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, ... 34 0.054
BP059197 34 0.071
CB828653 33 0.16
>TC18924 homologue to UP|Q94FN3 (Q94FN3) Phosphatidylinositol transfer-like
protein II, complete
Length = 1976
Score = 631 bits (1627), Expect = 0.0
Identities = 330/546 (60%), Positives = 403/546 (73%), Gaps = 8/546 (1%)
Frame = +3
Query: 7 EKKKKVGSIKKVALSASSKFKNSFTKKGRKHSRVMSICIEDSFDAEELQAVDALRQTLIL 66
+K K GS+KK A+S S K R S+VMS+ IED DAE+L+AVD RQ LIL
Sbjct: 75 DKSDKAGSLKKKAMSLRSSLSR---KSRRSSSKVMSVEIEDVRDAEDLKAVDEFRQALIL 245
Query: 67 EELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEVL 126
+ELLP KHDD H +LRFL+ARK+D EK+KQMW+DML+WRKEFGADTI+EDF+F+E+DEV+
Sbjct: 246 DELLPEKHDDYHQLLRFLKARKFDTEKSKQMWSDMLQWRKEFGADTIVEDFDFKEIDEVV 425
Query: 127 KCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACSI 186
K YP GHHGVDKDGRPVYIE +GQVD KL+QVT+++RY+KYHV+EFER F +K ACSI
Sbjct: 426 KYYPHGHHGVDKDGRPVYIENIGQVDATKLMQVTTMDRYIKYHVKEFERTFDLKFAACSI 605
Query: 187 AAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRL 246
AAKKHIDQSTTILDVQGVGL++ NK AR+L+ RLQKIDGDNYPE+LNRMFIINAGSGFR+
Sbjct: 606 AAKKHIDQSTTILDVQGVGLKNFNKHARELITRLQKIDGDNYPETLNRMFIINAGSGFRM 785
Query: 247 LWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCMLSDKGP 306
LW+TVKSFLDPKTTSKIHVLGNKYQSKLLEVIDAS+LPEFLGGTCTCAD+GGCM SDKGP
Sbjct: 786 LWSTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASQLPEFLGGTCTCADQGGCMRSDKGP 965
Query: 307 WNDPEILKMAQNGVGRYTIKALSG-VEEKTIKQEETAYQKGFKDSFPETLDVHCLDQPKS 365
W DPE+++M QNG + + K S VEEK I +E T F + +
Sbjct: 966 WKDPELVRMVQNGEHKCSRKCESPVVEEKKIPEETTKMGANFTSQLSSV-----FGEVPA 1130
Query: 366 YGVYQYDSFVPVLDKAVDSSWKKTIQNDKYALSKDCFSNNNGMNS----SGFSKQFVGGI 421
Y+ FVP D+ V WKK +N+++ +SK N +S + Q G+
Sbjct: 1131TKACNYEDFVPAADETV---WKKIEENEQFQMSKAAVQTFNMADSCKIHEKVNSQIFTGV 1301
Query: 422 MALVMGIVTIIRMTSSMPRKITEAALYGGNSVYYDG---SMIKAAAISNNEYMAMMKRMA 478
MA VMG VT++RMT +MP+K+T+A Y NSVY G S +IS E+M +MKRMA
Sbjct: 1302MAFVMGNVTMVRMTRNMPKKLTDANFY-SNSVYSGGQNPSDQTNPSISAQEFMTVMKRMA 1478
Query: 479 ELEEKVTVLSVKPVMPPEKEEMLNNALTRVSTLEQELGATKKALEDALTRQVELEGQIDK 538
+LEEK+ ++ K MPPEKEEMLN A++R LEQEL +TKKALED+L +Q EL I+K
Sbjct: 1479KLEEKMGNMNYKTCMPPEKEEMLNAAISRADALEQELMSTKKALEDSLAKQEELSAYIEK 1658
Query: 539 KKKKKK 544
KKKKKK
Sbjct: 1659KKKKKK 1676
>TC8616 UP|Q94FN1 (Q94FN1) Phosphatidylinositol transfer-like protein III,
complete
Length = 2302
Score = 357 bits (917), Expect(2) = e-154
Identities = 215/449 (47%), Positives = 274/449 (60%), Gaps = 44/449 (9%)
Frame = +3
Query: 145 IERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACSIAAKKHIDQSTTILDVQGV 204
+ERLG+VD NKL+QVT+++RY++YHV+EFE++FA+K PAC++AAK+HID STTILDVQGV
Sbjct: 603 LERLGKVDPNKLMQVTTMDRYVRYHVQEFEKSFAIKFPACTMAAKRHIDSSTTILDVQGV 782
Query: 205 GLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRLLWNTVKSFLDPKTTSKIH 264
GL++ K+AR+L+ RLQK+DGDNYPE+L +MFIINAG GFRLLWNTVKSFLDPKTTSKIH
Sbjct: 783 GLKNFTKSARELITRLQKVDGDNYPETLCQMFIINAGPGFRLLWNTVKSFLDPKTTSKIH 962
Query: 265 VLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCMLSDKGPWNDPEILKMAQNGV---G 321
VLGNKY SK LEVIDASELPE LGG CTC D+GGC+ SDKGPW +PEILKM NG
Sbjct: 963 VLGNKYHSKWLEVIDASELPECLGGACTCEDQGGCLRSDKGPWKNPEILKMVLNGEPRRA 1142
Query: 322 RYTIKALSGVEEKTIKQEETAYQ--KGFKDSFPET-LDVHCLDQPKSYGVYQ-------- 370
R +K L+ E K I + Y KG S E+ + + PK+ Y
Sbjct: 1143 RQVVKVLNS-EGKVIAYAKPRYPTVKGSDTSTAESGSEAEDIASPKAMKNYSHLRLTPVR 1319
Query: 371 -----------------YDSFVPVLDKAVDSSWKKTIQNDKYALSKDCFSNNNGMNS-SG 412
YD +VP++DK VD+ KK + S+ S + G
Sbjct: 1320 EEAKIVGKSSYTNNFSGYDEYVPMVDKPVDAVLKKQASLQRSYTSQGAPSRPATQRTPEG 1499
Query: 413 FSKQFVGGIMALVMGIVTII-----RMTSSMPR------KITEAALYGGNSVYYDGSMIK 461
+ + I A ++ I TI R+T +P + T + V S
Sbjct: 1500 IQARILVAITAFLLTIFTIFRQVACRVTKKLPAISSNHDQSTSEPTFDTTVVEVIPSSST 1679
Query: 462 AAAISNNEYMAMMKRMAELEEKVTVLSVKP-VMPPEKEEMLNNALTRVSTLEQELGATKK 520
A N +M+KR+ ELEEKV L KP MP EKEE+LN A+ RV LE EL ATKK
Sbjct: 1680 PAHTEENLLPSMLKRLGELEEKVDTLQSKPSEMPYEKEELLNAAVCRVDALEAELIATKK 1859
Query: 521 ALEDALTRQVELEGQIDKKKKKKKKLVRY 549
AL +AL RQ EL ID++ + K + R+
Sbjct: 1860 ALYEALMRQEELLAYIDRQAEAKLRKKRF 1946
Score = 206 bits (524), Expect(2) = e-154
Identities = 99/147 (67%), Positives = 121/147 (81%), Gaps = 3/147 (2%)
Frame = +2
Query: 2 EHSEDEKKKKVGSIKKVALSASSKFKNSFTKKGRKHS---RVMSICIEDSFDAEELQAVD 58
E+SEDE++ ++GS+KK ALSAS+KFK+S KK + RV S+ IED D EELQAVD
Sbjct: 164 ENSEDERRTRIGSLKKKALSASTKFKHSLRKKSSRRKSDGRVSSVSIEDVRDVEELQAVD 343
Query: 59 ALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFE 118
A RQ+LI++ELLP DD HMMLRFL+ARK+DIEK K MW +ML+WRKEFGADTIM+DFE
Sbjct: 344 AFRQSLIMDELLPQAFDDYHMMLRFLKARKFDIEKAKHMWAEMLQWRKEFGADTIMQDFE 523
Query: 119 FEELDEVLKCYPQGHHGVDKDGRPVYI 145
F+ELDEV++ YP GHHGVDK+GRPVYI
Sbjct: 524 FQELDEVVRYYPHGHHGVDKEGRPVYI 604
>TC14782 UP|O04360 (O04360) Late nodulin Nlj16, complete
Length = 1010
Score = 88.6 bits (218), Expect = 2e-18
Identities = 45/88 (51%), Positives = 63/88 (71%), Gaps = 1/88 (1%)
Frame = +3
Query: 466 SNNEYMAMMKRMAELEEKVTVLSVKPV-MPPEKEEMLNNALTRVSTLEQELGATKKALED 524
++ E+ +MKRMAELEEK+T ++ +P MPPEKEEMLN ++R LE++L TKKALED
Sbjct: 210 TSKEFTTVMKRMAELEEKMTTMNHQPATMPPEKEEMLNATISRADVLEKQLMDTKKALED 389
Query: 525 ALTRQVELEGQIDKKKKKKKKLVRYICC 552
+L +Q L ++KKK+KKK + CC
Sbjct: 390 SLAKQEVLSAYVEKKKQKKK---TFFCC 464
>BP039806
Length = 546
Score = 55.1 bits (131), Expect(2) = 2e-17
Identities = 26/38 (68%), Positives = 33/38 (86%)
Frame = -3
Query: 450 GNSVYYDGSMIKAAAISNNEYMAMMKRMAELEEKVTVL 487
G+SVYYDG+++KA IS +E+MA+MKRMAELEEKV L
Sbjct: 286 GSSVYYDGNIMKAPEISIDEHMALMKRMAELEEKVNAL 173
Score = 50.8 bits (120), Expect(2) = 2e-17
Identities = 23/49 (46%), Positives = 33/49 (66%)
Frame = -2
Query: 400 DCFSNNNGMNSSGFSKQFVGGIMALVMGIVTIIRMTSSMPRKITEAALY 448
DCF N + ++G + QF GGI + MG +T IR+T +MPRKITE ++
Sbjct: 425 DCFPNKDC--NTGLTTQFAGGIKVVAMGFITFIRLTRNMPRKITEVGVW 285
>TC14783 UP|O04360 (O04360) Late nodulin Nlj16, complete
Length = 540
Score = 62.4 bits (150), Expect(2) = 7e-14
Identities = 31/58 (53%), Positives = 43/58 (73%), Gaps = 1/58 (1%)
Frame = +2
Query: 466 SNNEYMAMMKRMAELEEKVTVLSVKPV-MPPEKEEMLNNALTRVSTLEQELGATKKAL 522
++ E+ +MKRMAELEEK+T ++ +P MPPEKEEMLN ++R LE++L TKK L
Sbjct: 182 TSKEFTTVMKRMAELEEKMTTMNHQPATMPPEKEEMLNATISRADVLEKQLMDTKKVL 355
Score = 31.6 bits (70), Expect(2) = 7e-14
Identities = 15/33 (45%), Positives = 21/33 (63%)
Frame = +1
Query: 520 KALEDALTRQVELEGQIDKKKKKKKKLVRYICC 552
+ALED+L +Q L + KKK+KKK + CC
Sbjct: 400 QALEDSLAKQEVLSAYVXKKKQKKK---TFFCC 489
>TC14753 similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial (57%)
Length = 1472
Score = 73.6 bits (179), Expect = 8e-14
Identities = 59/217 (27%), Positives = 102/217 (46%), Gaps = 6/217 (2%)
Frame = +1
Query: 79 MMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEVLKCYPQGHHGVDK 138
++L+FLRAR + ++ M + ++WRKEFG D ++++ E D+V+ + QGH DK
Sbjct: 88 VLLKFLRARDFKVKDAFTMIKNTVRWRKEFGIDALVDEDLGSEWDKVV--FTQGH---DK 252
Query: 139 DGRPVYIERLGQVDCNKLLQVT-----SVERYLKYHVREFERAFAVKLPACSIAAKKHID 193
G V G+ + +L Q T ++++K+ ++ E++ + + A I
Sbjct: 253 LGHAVCYNVFGEFEDKELYQKTFSDDEKRQKFIKWRIQVLEKSVRKLDFSPTPTAISTIV 432
Query: 194 QSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRLLWNTVKS 253
Q + + G+G + +A LQ LQ DNYPE + + IN + +
Sbjct: 433 QVNDLKNSPGLGKWELRQATNQALQLLQ----DNYPEFVAKQVFINVPWWYLAFSRMISP 600
Query: 254 FLDPKTTSKIHVLG-NKYQSKLLEVIDASELPEFLGG 289
FL +T SK G +K L + I +P GG
Sbjct: 601 FLTQRTKSKFVFAGPSKSAETLFKYIAPELVPVQYGG 711
>AV769442
Length = 559
Score = 72.8 bits (177), Expect = 1e-13
Identities = 41/70 (58%), Positives = 52/70 (73%), Gaps = 1/70 (1%)
Frame = -2
Query: 475 KRMAELEEKVTVLSVKP-VMPPEKEEMLNNALTRVSTLEQELGATKKALEDALTRQVELE 533
KR+ ELEEKV +L+ KP +MP EKEE+LN A+ RV LE EL ATKKAL +AL RQ EL
Sbjct: 465 KRLCELEEKVDILNSKPNIMPYEKEELLNAAVYRVDALEAELIATKKALYEALIRQEELL 286
Query: 534 GQIDKKKKKK 543
ID ++++K
Sbjct: 285 AYIDCQERRK 256
>AV409080
Length = 423
Score = 61.6 bits (148), Expect = 3e-10
Identities = 33/63 (52%), Positives = 45/63 (71%), Gaps = 3/63 (4%)
Frame = +2
Query: 2 EHSEDEKKKKVGSIKKVALSASSKFKNSFTKKGRKH---SRVMSICIEDSFDAEELQAVD 58
E SEDE++ ++GS+KK A++ASSKF++S KK + SR S+ IED D +ELQAVD
Sbjct: 233 EVSEDERRTRIGSLKKKAINASSKFRHSLRKKSSRRKNASRSNSVSIEDIRDVKELQAVD 412
Query: 59 ALR 61
A R
Sbjct: 413 AFR 421
>AV428489
Length = 386
Score = 58.9 bits (141), Expect = 2e-09
Identities = 31/96 (32%), Positives = 50/96 (51%)
Frame = +1
Query: 191 HIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRLLWNT 250
+I +LD+ G+ ++N+ LL + +D NYPE + +I+NA F W
Sbjct: 1 YIGTCVKVLDMSGLKFSALNQLR--LLTAISTVDDLNYPEKTDTYYIVNAPYVFSACWKV 174
Query: 251 VKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEF 286
VK L +T KI VL + +LL+++D + LP F
Sbjct: 175 VKPLLQERTRRKIQVLQGSGKEELLKIMDYASLPHF 282
>BP064990
Length = 470
Score = 55.1 bits (131), Expect = 3e-08
Identities = 27/47 (57%), Positives = 37/47 (78%)
Frame = +1
Query: 12 VGSIKKVALSASSKFKNSFTKKGRKHSRVMSICIEDSFDAEELQAVD 58
V S KK A++AS+ +NS T+KGR+ S+VMS+ IED DAEEL+AV+
Sbjct: 322 VDSFKKKAINASNMLRNSLTRKGRRSSKVMSVEIEDVHDAEELKAVE 462
>TC16928 UP|Q94FN0 (Q94FN0) Phosphatidylinositol transfer-like protein IV,
partial (12%)
Length = 442
Score = 48.9 bits (115), Expect = 2e-06
Identities = 21/24 (87%), Positives = 24/24 (99%)
Frame = -2
Query: 230 ESLNRMFIINAGSGFRLLWNTVKS 253
++LNRMFIINAGSGFR+LWNTVKS
Sbjct: 99 QTLNRMFIINAGSGFRILWNTVKS 28
Score = 44.7 bits (104), Expect = 4e-05
Identities = 24/47 (51%), Positives = 29/47 (61%)
Frame = -3
Query: 205 GLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRLLWNTV 251
GL+S NK AR+L+ R+QK+DGDNYPE I G F L N V
Sbjct: 278 GLKSFNKHARELVTRIQKVDGDNYPEVCFSTLIKLWGQEFGL*INIV 138
>TC17702 weakly similar to GB|AAN73306.1|25141223|BT002309
At3g51670/T18N14_50 {Arabidopsis thaliana;}, partial
(20%)
Length = 574
Score = 48.5 bits (114), Expect = 3e-06
Identities = 27/66 (40%), Positives = 39/66 (58%), Gaps = 1/66 (1%)
Frame = +2
Query: 79 MMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIM-EDFEFEELDEVLKCYPQGHHGVD 137
++L+FLRAR + + M L WRKEFGAD I+ E+ F+EL+ V+ C D
Sbjct: 374 LLLKFLRARDFRVCDALNMLLKCLAWRKEFGADNIVDEELGFKELEGVVAC----TQVYD 541
Query: 138 KDGRPV 143
++G PV
Sbjct: 542 REGHPV 559
>TC11997 similar to UP|O48940 (O48940) Polyphosphoinositide binding protein
Ssh2p, partial (26%)
Length = 492
Score = 45.1 bits (105), Expect = 3e-05
Identities = 25/57 (43%), Positives = 34/57 (58%), Gaps = 1/57 (1%)
Frame = +2
Query: 234 RMFIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGN-KYQSKLLEVIDASELPEFLGG 289
++FI++A F +W V F+D T KI + N K +S LLE ID S+LPE GG
Sbjct: 2 KLFIVHAPYIFMKVWKLVYPFIDDHTKKKIVFVDNQKLKSTLLEEIDESQLPEIYGG 172
>TC14873 weakly similar to GB|BAB08982.1|9758453|AB008271 selenium-binding
protein-like {Arabidopsis thaliana;} , partial (7%)
Length = 763
Score = 44.3 bits (103), Expect = 5e-05
Identities = 30/83 (36%), Positives = 42/83 (50%), Gaps = 3/83 (3%)
Frame = +2
Query: 213 ARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRLLWNTVKSFLDPKTTSKIHVL--GNKY 270
ARD++ L +YPE L F+ N S F+ + VK FLDPKT K+ + NK
Sbjct: 50 ARDIIHILLY----HYPERLAIAFLFNPPSIFQAFYKAVKYFLDPKTAQKVKFVYPNNKD 217
Query: 271 QSKLLE-VIDASELPEFLGGTCT 292
+L++ + D LP GG T
Sbjct: 218 SVELMKSLFDIDNLPSEFGGKAT 286
>AW720085
Length = 524
Score = 38.1 bits (87), Expect = 0.004
Identities = 28/98 (28%), Positives = 47/98 (47%), Gaps = 6/98 (6%)
Frame = +1
Query: 80 MLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIM------EDFEFEELDEVLKCYPQGH 133
++RFL+AR+++ K +M D L WR + D+I+ ED D L G
Sbjct: 235 LMRFLKAREWNASKAHKMLVDCLNWRVQSEIDSILSKPIIPEDLYRAVRDSQL----IGL 402
Query: 134 HGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVR 171
G ++G PV+ +G +K SV Y++ H++
Sbjct: 403 SGFSREGLPVFAIGVGLSTFDK----ASVHYYVQSHIQ 504
>CB828378
Length = 554
Score = 37.7 bits (86), Expect = 0.005
Identities = 17/28 (60%), Positives = 21/28 (74%)
Frame = +3
Query: 493 MPPEKEEMLNNALTRVSTLEQELGATKK 520
MPPEKEEMLN ++R LE++L TKK
Sbjct: 18 MPPEKEEMLNATISRADVLEKQLMDTKK 101
Score = 31.6 bits (70), Expect = 0.35
Identities = 14/28 (50%), Positives = 20/28 (71%)
Frame = +1
Query: 520 KALEDALTRQVELEGQIDKKKKKKKKLV 547
+ALED+L +Q L ++KKK+KKK V
Sbjct: 241 QALEDSLAKQEVLSAYVEKKKQKKKTFV 324
>TC13447
Length = 554
Score = 37.4 bits (85), Expect = 0.006
Identities = 21/71 (29%), Positives = 37/71 (51%)
Frame = +2
Query: 75 DDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEVLKCYPQGHH 134
DD M+ FL+ RK+ +++ T +KWR++F + E+ ++ K Y H
Sbjct: 173 DDEDMIQWFLKDRKFSVDEAVSKLTKAIKWRQDFEVSKLTEE-AVKDAARSGKAYV--HD 343
Query: 135 GVDKDGRPVYI 145
+D +GRPV +
Sbjct: 344 FLDINGRPVLV 376
>TC13909 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial
(20%)
Length = 399
Score = 34.3 bits (77), Expect = 0.054
Identities = 21/63 (33%), Positives = 36/63 (56%), Gaps = 7/63 (11%)
Frame = +3
Query: 53 ELQAVDALRQTLILEEL-----LPSKHDDPH--MMLRFLRARKYDIEKTKQMWTDMLKWR 105
E +AV L+Q L + + +P DD ++L+FLRAR++ ++ M + L+WR
Sbjct: 204 ETKAVQDLKQLLTVTDEPSIWGVPLLKDDRTDVILLKFLRAREFKVKDALLMINNTLRWR 383
Query: 106 KEF 108
K+F
Sbjct: 384 KDF 392
>BP059197
Length = 365
Score = 33.9 bits (76), Expect = 0.071
Identities = 14/30 (46%), Positives = 22/30 (72%)
Frame = +3
Query: 466 SNNEYMAMMKRMAELEEKVTVLSVKPVMPP 495
++ E+ +MKRMAELEEK+T ++ +P P
Sbjct: 276 TSKEFTTVMKRMAELEEKMTTMNHQPATMP 365
>CB828653
Length = 524
Score = 32.7 bits (73), Expect = 0.16
Identities = 15/43 (34%), Positives = 27/43 (61%)
Frame = +1
Query: 501 LNNALTRVSTLEQELGATKKALEDALTRQVELEGQIDKKKKKK 543
L +L+R+ ++E +L TKKAL ++QVEL ++ K+ +
Sbjct: 1 LQESLSRIKSIEYDLQKTKKALLATASKQVELAESLESLKESR 129
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,307,231
Number of Sequences: 28460
Number of extensions: 99680
Number of successful extensions: 705
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 629
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 680
length of query: 555
length of database: 4,897,600
effective HSP length: 95
effective length of query: 460
effective length of database: 2,193,900
effective search space: 1009194000
effective search space used: 1009194000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC140722.2


