

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.8 + phase: 0 /pseudo
(59 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
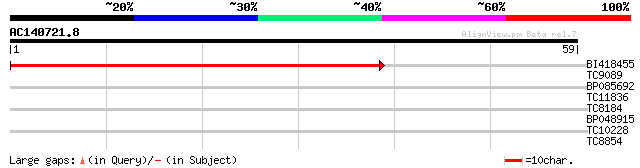
Score E
Sequences producing significant alignments: (bits) Value
BI418455 62 2e-11
TC9089 similar to UP|RHRE_PEA (Q9S8P4) Rhicadhesin receptor prec... 30 0.073
BP085692 29 0.21
TC11836 similar to GB|AAB51577.1|1755176|ATU75199 germin-like pr... 28 0.36
TC8184 similar to UP|Q9LJQ4 (Q9LJQ4) Muconate cycloisomerase-lik... 25 2.3
BP048915 24 5.2
TC10228 similar to UP|Q9LDF5 (Q9LDF5) 3-hydroxybutyryl-CoA dehyd... 24 6.8
TC8854 weakly similar to UP|Q9LEU4 (Q9LEU4) CCR4-associated fact... 23 8.9
>BI418455
Length = 477
Score = 62.4 bits (150), Expect = 2e-11
Identities = 29/39 (74%), Positives = 33/39 (84%)
Frame = +1
Query: 1 MKILYLLVSILALASSIAFAYDPSPLQDFCVAIKDPKDG 39
M+++Y LV+ILALASS A AYDPSPLQDFCV I DPK G
Sbjct: 16 MEVVYFLVTILALASSFASAYDPSPLQDFCVGINDPKYG 132
Score = 28.9 bits (63), Expect = 0.21
Identities = 13/20 (65%), Positives = 15/20 (75%)
Frame = +2
Query: 40 DYSDLTHLVYLWLALILPLR 59
+Y D THL YLWLA IL L+
Sbjct: 260 NYLD*THLAYLWLA*ILHLK 319
>TC9089 similar to UP|RHRE_PEA (Q9S8P4) Rhicadhesin receptor precursor
(Germin-like protein), partial (93%)
Length = 981
Score = 30.4 bits (67), Expect = 0.073
Identities = 18/36 (50%), Positives = 20/36 (55%)
Frame = +1
Query: 4 LYLLVSILALASSIAFAYDPSPLQDFCVAIKDPKDG 39
L L LAL + AF+ DP LQD CVA DP G
Sbjct: 172 LLLFALPLALLLTTAFSSDPDYLQDLCVA--DPASG 273
>BP085692
Length = 334
Score = 28.9 bits (63), Expect = 0.21
Identities = 13/34 (38%), Positives = 22/34 (64%)
Frame = -2
Query: 1 MKILYLLVSILALASSIAFAYDPSPLQDFCVAIK 34
M++LYL++ ++ L SIAF +D + FC+ K
Sbjct: 333 MRLLYLIMFLITLVISIAF*FDCLCVGSFCLGDK 232
>TC11836 similar to GB|AAB51577.1|1755176|ATU75199 germin-like protein
{Arabidopsis thaliana;} , partial (56%)
Length = 434
Score = 28.1 bits (61), Expect = 0.36
Identities = 13/32 (40%), Positives = 19/32 (58%)
Frame = +3
Query: 1 MKILYLLVSILALASSIAFAYDPSPLQDFCVA 32
+ I+ L+V +L ++ A DP LQD CVA
Sbjct: 45 LHIMKLVVLLLLTLTTFTAASDPDSLQDICVA 140
>TC8184 similar to UP|Q9LJQ4 (Q9LJQ4) Muconate cycloisomerase-like protein,
partial (17%)
Length = 683
Score = 25.4 bits (54), Expect = 2.3
Identities = 12/32 (37%), Positives = 19/32 (58%), Gaps = 6/32 (18%)
Frame = -1
Query: 32 AIKDPKDGD------YSDLTHLVYLWLALILP 57
A+K+ K + YS + ++YLWL L+LP
Sbjct: 500 AVKEAKHSNCQMLASYSCCSRILYLWLLLLLP 405
>BP048915
Length = 562
Score = 24.3 bits (51), Expect = 5.2
Identities = 13/35 (37%), Positives = 21/35 (59%)
Frame = +1
Query: 15 SSIAFAYDPSPLQDFCVAIKDPKDGDYSDLTHLVY 49
S I AY+ + + D C IKD + DY ++++VY
Sbjct: 31 SLIVLAYEDNKIHDKCYIIKD-RQTDY--ISYIVY 126
>TC10228 similar to UP|Q9LDF5 (Q9LDF5) 3-hydroxybutyryl-CoA
dehydrogenase-like protein, partial (44%)
Length = 572
Score = 23.9 bits (50), Expect = 6.8
Identities = 13/29 (44%), Positives = 17/29 (57%), Gaps = 1/29 (3%)
Frame = -3
Query: 25 PLQDFCVAIKDPKDGDYSDLTHLVY-LWL 52
P+Q +A+KDP DG Y + Y LWL
Sbjct: 255 PIQ*SQLALKDPLDG*YPASSRYQYPLWL 169
>TC8854 weakly similar to UP|Q9LEU4 (Q9LEU4) CCR4-associated factor-like
protein, partial (8%)
Length = 916
Score = 23.5 bits (49), Expect = 8.9
Identities = 7/12 (58%), Positives = 10/12 (83%)
Frame = +2
Query: 46 HLVYLWLALILP 57
HL+YLWL+ + P
Sbjct: 128 HLLYLWLSFLFP 163
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.328 0.145 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,260,928
Number of Sequences: 28460
Number of extensions: 13701
Number of successful extensions: 93
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 93
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 93
length of query: 59
length of database: 4,897,600
effective HSP length: 35
effective length of query: 24
effective length of database: 3,901,500
effective search space: 93636000
effective search space used: 93636000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 49 (23.5 bits)
Medicago: description of AC140721.8


