

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140671.7 - phase: 0
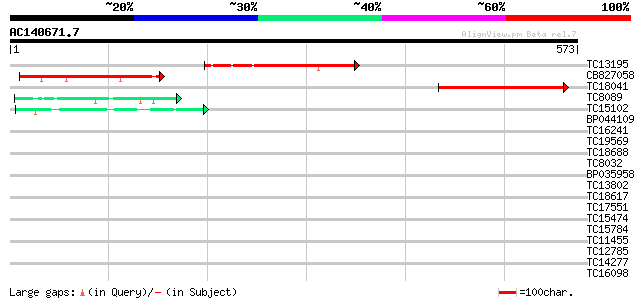
(573 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC13195 182 2e-46
CB827058 166 1e-41
TC18041 weakly similar to UP|O80588 (O80588) At2g44190 protein, ... 106 1e-23
TC8089 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinog... 48 5e-06
TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, ... 42 4e-04
BP044109 40 0.001
TC16241 weakly similar to UP|Q91TH4 (Q91TH4) T122, partial (7%) 37 0.007
TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor k... 37 0.009
TC18688 weakly similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (13%) 37 0.009
TC8032 similar to UP|Q9M0B6 (Q9M0B6) Nucleotide sugar epimerase-... 37 0.009
BP035958 37 0.009
TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-... 37 0.011
TC18617 weakly similar to UP|Q9FPQ5 (Q9FPQ5) Gamete-specific hyd... 36 0.015
TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, part... 35 0.033
TC15474 UP|Q7Y1B9 (Q7Y1B9) Ammonium transporter, complete 34 0.056
TC15784 similar to UP|PE31_ARATH (Q9LHA7) Peroxidase 31 precurso... 34 0.074
TC11455 similar to UP|Q9ZUS8 (Q9ZUS8) At2g37380 protein, partial... 34 0.074
TC12785 similar to UP|Q7X9B2 (Q7X9B2) 9/13 hydroperoxide lyase, ... 34 0.074
TC14277 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (AT1G65720/F1E22_1... 34 0.074
TC16098 homologue to UP|Q8LAQ5 (Q8LAQ5) Flower pigmentation prot... 33 0.13
>TC13195
Length = 482
Score = 182 bits (461), Expect = 2e-46
Identities = 107/163 (65%), Positives = 121/163 (73%), Gaps = 7/163 (4%)
Frame = +1
Query: 198 PAKSQQNQANFMNRSLDCGVLLRSSNGFGSNVVRSL-RNSLLDPRVSHDGATLRSESNKN 256
P + QQN ANFMNRSLDC +R NG NVVRSL +NS+ D R S D +TL SE NKN
Sbjct: 1 PTRPQQN-ANFMNRSLDCTDSVRKLNG-SRNVVRSLLQNSMADVRASQD-STLSSEINKN 171
Query: 257 GGSEPVIEPELVPSDNESVTSGSSSGVLDYGGGKPQR-SARAIVVPARFLQEATN----- 310
GGSEP EPE VPSDNESVTSGSSSG ++GGG+ QR S+RAIVVPARF QEA N
Sbjct: 172 GGSEPESEPEPVPSDNESVTSGSSSGAQEWGGGQAQRASSRAIVVPARFWQEANNRLRHQ 351
Query: 311 PSSRNGGIGNRSTVPPKLLVPKKSVFDSPASSPRGIVNNRLQG 353
+ G GN++TVPPKLL PKKS FDSP SSPRG+VN+RLQG
Sbjct: 352 TEPQPSGNGNKATVPPKLLAPKKSGFDSPVSSPRGVVNSRLQG 480
>CB827058
Length = 555
Score = 166 bits (419), Expect = 1e-41
Identities = 102/162 (62%), Positives = 113/162 (68%), Gaps = 16/162 (9%)
Frame = +1
Query: 11 TKNTKRAPTPTRPPLLPSES---DNAIVRKPKAREVTSRYMYSSSSSSS---FSSPPRRT 64
T TKRAPTPTRPPLLPS + + R+ K+REVTSRYM SSSSSSS SSPPRR
Sbjct: 79 TTTTKRAPTPTRPPLLPSGNVAPPPPLSRRVKSREVTSRYMSSSSSSSSSSSVSSPPRRY 258
Query: 65 NSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQ--RQGTPRPNG--------ETPV 114
+SP+V RTVNS+KQKS+ P VKRSQS ERQRQ RQGTPRP+G + P
Sbjct: 259 HSPIVPRTVNSSKQKSSTIAVPTPQVKRSQSAERQRQRPRQGTPRPSGAGVVASAADAPA 438
Query: 115 AQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRS 156
A KMLFTS RSLSVSFQGESFSI VSK KP QS+R S
Sbjct: 439 AHKMLFTSTRSLSVSFQGESFSIQVSKPKP---APVQSVRNS 555
Score = 26.9 bits (58), Expect = 9.0
Identities = 12/18 (66%), Positives = 13/18 (71%)
Frame = +1
Query: 152 SLRRSTPERRKVPATVTT 169
S+RRSTPERR T TT
Sbjct: 37 SMRRSTPERRLAAETTTT 90
>TC18041 weakly similar to UP|O80588 (O80588) At2g44190 protein, partial
(11%)
Length = 925
Score = 106 bits (264), Expect = 1e-23
Identities = 56/131 (42%), Positives = 90/131 (67%)
Frame = +3
Query: 434 MQWRFVNARADASLSVQTLNSEKSLYAAWVATSKLRESVVAKRIMLQLLKQHLKLISILN 493
+QWRF NA+A AS+ Q SEK+LY+ + S++R+SV KR L+LL+++ L +IL+
Sbjct: 3 LQWRFANAKAAASMKAQQKESEKALYSHAMKISEMRDSVNRKRAELELLQRYKTLSTILD 182
Query: 494 EQMIYLEDWAILDRVYSGSLSGATEALKASTLRLPVFGGAKIDLLNLKEAICSAMDVMQA 553
QM YLE+W+ ++ YS SL+ AT+AL ++++LPV G ++D + EA+ SA +M+
Sbjct: 183 AQMPYLEEWSSMEEDYSVSLTEATQALVNASVQLPVGGNVRVDEREVGEALNSASKMMET 362
Query: 554 MASSICLLLPK 564
+ S+I L+PK
Sbjct: 363 IVSNIQRLVPK 395
>TC8089 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (43%)
Length = 890
Score = 47.8 bits (112), Expect = 5e-06
Identities = 54/200 (27%), Positives = 78/200 (39%), Gaps = 32/200 (16%)
Frame = +2
Query: 6 STTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTN 65
ST ++ +T +P+PT PP S N + P A T +++SS ++ SPP RT+
Sbjct: 254 STPSSPPSTTTSPSPTSPP----RSTNG-PQSPSAPSTTP--PWTTSSQNTHQSPPSRTS 412
Query: 66 SPLVNRTVNSTKQKSTPSVT------------------------PAAFVKRSQSTERQRQ 101
SP + + S + ST S T P + +S S R
Sbjct: 413 SPSTSSSTTSAPRSSTRSPTAPPSPPQCTKPPAPPRAPPDSSTSPTSAAGKSDSAPRTTT 592
Query: 102 RQGTPRPNGETPVAQKMLFTSARSLSVSF-----QGESFSIPVSKAKP---PSATVTQSL 153
P P + +K TS S SV F Q P+S+ P PS S
Sbjct: 593 ---APSPLRSSNP*RKSPTTSPSSRSVRFFPPPQQKPLHPHPLSRISPPLCPSTVARSSP 763
Query: 154 RRSTPERRKVPATVTTPTIG 173
S P R + + TT T+G
Sbjct: 764 TLSPPRRTRTQPSPTTSTVG 823
Score = 35.4 bits (80), Expect = 0.025
Identities = 38/154 (24%), Positives = 58/154 (36%)
Frame = +2
Query: 23 PPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVNRTVNSTKQKSTP 82
P LLP ++ P T S+++S S +SPPR TN P + S P
Sbjct: 203 PWLLPPQTPTT---SPAFSPSTPSSPPSTTTSPSPTSPPRSTNGP---------QSPSAP 346
Query: 83 SVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQGESFSIPVSKA 142
S TP + + R +P + T A + +S RS P +
Sbjct: 347 STTPPWTTSSQNTHQSPPSRTSSPSTSSSTTSAPR---SSTRS------------PTAPP 481
Query: 143 KPPSATVTQSLRRSTPERRKVPATVTTPTIGRGR 176
PP T + R+ P+ + T+PT G+
Sbjct: 482 SPPQCTKPPAPPRAPPD------SSTSPTSAAGK 565
>TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, partial
(68%)
Length = 1097
Score = 41.6 bits (96), Expect = 4e-04
Identities = 48/201 (23%), Positives = 75/201 (36%), Gaps = 6/201 (2%)
Frame = +1
Query: 7 TTTTTKNTKRAPTPTRPP----LLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
TT TT +T +A T + PP P S+ + P + + SS S + PP
Sbjct: 175 TTATTTSTTKAATNSSPPESRSTTPPSSEPTCLLAPASPP-------TPKSSHSSTPPPS 333
Query: 63 RTNS--PLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLF 120
R++S P + T P +PAA T P N P + K
Sbjct: 334 RSSSTTPKSKNATSLTAPSCPPPASPAATAPAPSKTPPN------PPTNSSKPSSCKSSL 495
Query: 121 TSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGN 180
+ S+S + PPS T + S+PE ++ PA+ T P+ + +
Sbjct: 496 PNTTPSSLS------------SAPPSPTTSPISTPSSPENQEKPAS-TPPSAPPRSTSSS 636
Query: 181 SDQTENSISRSGDQHRWPAKS 201
S N+ + + PA S
Sbjct: 637 SSSPINTPRKPSSATKAPASS 699
Score = 30.0 bits (66), Expect = 1.1
Identities = 20/65 (30%), Positives = 35/65 (53%), Gaps = 1/65 (1%)
Frame = +1
Query: 3 TAISTTTTTKNTKRAPTPTRPPL-LPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPP 61
+++ TT + + P+PT P+ PS +N KP + ++ +SSSSSS + P
Sbjct: 487 SSLPNTTPSSLSSAPPSPTTSPISTPSSPENQ--EKPASTPPSAPPRSTSSSSSSPINTP 660
Query: 62 RRTNS 66
R+ +S
Sbjct: 661 RKPSS 675
>BP044109
Length = 497
Score = 39.7 bits (91), Expect = 0.001
Identities = 37/122 (30%), Positives = 53/122 (43%)
Frame = +2
Query: 6 STTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTN 65
++ T+K+T R TPT PP P+ S PK T + S+S +P ++
Sbjct: 161 ASNATSKSTPRTQTPTTPPPSPTSS-------PKPPPST---FATKPSTSPPETPSSSSH 310
Query: 66 SPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARS 125
P T+ S STP+ TP+ + ST R TP P P A++M S S
Sbjct: 311 GPA--PTLPSPPSSSTPTSTPSPPSPPNGSTLPSPPR-ATPTPAPSLPAARRMTSASLCS 481
Query: 126 LS 127
S
Sbjct: 482 TS 487
Score = 30.4 bits (67), Expect = 0.81
Identities = 35/122 (28%), Positives = 47/122 (37%)
Frame = +2
Query: 50 SSSSSSSFSSPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPN 109
S SSS F SPP P N+T S +T TP + T +P+P
Sbjct: 80 SLSSSHFFPSPPPPPPPPNKNKTTPSHASNATSKSTP----RTQTPTTPPPSPTSSPKPP 247
Query: 110 GETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTT 169
T A K + + S S G + ++P PPS+ S STP P T
Sbjct: 248 PST-FATKPSTSPPETPSSSSHGPAPTLP----SPPSS----STPTSTPSPPSPPNGSTL 400
Query: 170 PT 171
P+
Sbjct: 401 PS 406
>TC16241 weakly similar to UP|Q91TH4 (Q91TH4) T122, partial (7%)
Length = 799
Score = 37.4 bits (85), Expect = 0.007
Identities = 37/146 (25%), Positives = 63/146 (42%), Gaps = 13/146 (8%)
Frame = +1
Query: 6 STTTTTKNTKRAPTPT-RPPLL--PSESDNAIVRKPKA---REVTSRYMYSSSSSSSFSS 59
S T+ P+P+ +PP L P +S++ + P A R VT + S SSS + +S
Sbjct: 103 SQPPNTQTPANTPSPSSQPPYLSTPPKSNSPTSQPPVAQTPRAVTP--ISSPSSSPTSNS 276
Query: 60 PPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQG-------TPRPNGET 112
P TN P + +++++ TP+ TP S + G TP P+ T
Sbjct: 277 PYPSTNPPSLAPSISTSPPALTPASTPVPATASPPSQSPSAESPGASLPAATTPSPSSTT 456
Query: 113 PVAQKMLFTSARSLSVSFQGESFSIP 138
P + + ++ + G S + P
Sbjct: 457 PRSTQTPSSNNETTPAQRNGASSATP 534
>TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor kinase 1;1,
partial (16%)
Length = 486
Score = 37.0 bits (84), Expect = 0.009
Identities = 36/130 (27%), Positives = 57/130 (43%), Gaps = 8/130 (6%)
Frame = +1
Query: 50 SSSSSSSFSSPP----RRTNSPLVNRTVNSTKQKSTPSVT---PAAFVKRSQSTERQRQR 102
SSSSSSS+S PP + T+SP + T +T S VT P A ++S
Sbjct: 46 SSSSSSSYSPPPPPSQQPTHSPSTSPTSTTTPPPSPTKVTENPPTAPSTSTKSPTSSASE 225
Query: 103 QGTPRPNGETPVAQKMLFT-SARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERR 161
+ +P PN T Q+ + ++ S S S P + A P ++ ++ + TP
Sbjct: 226 EPSP-PNPSTSTTQQPKHSPTSPPASPSPSPPSTKPPTAMASPSTSPLSATKSHPTPPAA 402
Query: 162 KVPATVTTPT 171
++ PT
Sbjct: 403 PSASSTPPPT 432
>TC18688 weakly similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (13%)
Length = 514
Score = 37.0 bits (84), Expect = 0.009
Identities = 41/129 (31%), Positives = 51/129 (38%), Gaps = 9/129 (6%)
Frame = +3
Query: 39 KAREVTSRYMYSSSSSSSFS----SPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQ 94
+ ++S SSSSSS S SPP S L + TP P A S
Sbjct: 54 ECESLSSSSSSSSSSSSPLSLNQISPPNAPPS*LSAPPSPEERFSGTPPPPPPATGSAST 233
Query: 95 STERQRQRQGTPRPNGETPVAQKMLF-----TSARSLSVSFQGESFSIPVSKAKPPSATV 149
+T + P +P M TSA S+SVS + S P S P SAT
Sbjct: 234 ATPTPPTSSRSASPPSPSPANSHMASSPPSPTSALSVSVSTPSPAHSPPTSPPAPRSATS 413
Query: 150 TQSLRRSTP 158
T S R S+P
Sbjct: 414 TSS-RISSP 437
>TC8032 similar to UP|Q9M0B6 (Q9M0B6) Nucleotide sugar epimerase-like
protein, partial (97%)
Length = 2230
Score = 37.0 bits (84), Expect = 0.009
Identities = 45/159 (28%), Positives = 58/159 (36%), Gaps = 14/159 (8%)
Frame = +1
Query: 6 STTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTN 65
STTTTT K+ P+ PP S S A P +S + S S +S P T
Sbjct: 523 STTTTTHR*KKPAKPSSPPTAFS-SWKATSTMPSFSPSSSTWWRSPMSCTSPRRPESATP 699
Query: 66 SPLVNRTVNSTKQKSTPSVTPAAFVKRSQ------------STERQRQRQ--GTPRPNGE 111
++ T +T S+PS PA R+ ST R R R G P
Sbjct: 700 WRILIPTFTATSPPSSPSWKPANLQTRNPLLSGLLQAPSTVSTRRFRSRNPIGLTNPPAS 879
Query: 112 TPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVT 150
TP +K S ++S S S P A T
Sbjct: 880 TPRRRKPARKSPTPTTISTDSPSPG*DSSPFTDPGADPT 996
>BP035958
Length = 574
Score = 37.0 bits (84), Expect = 0.009
Identities = 41/156 (26%), Positives = 63/156 (40%), Gaps = 6/156 (3%)
Frame = +1
Query: 50 SSSSSSSFSSPPRRTNSPL------VNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQ 103
SSSSSSS PPRR +SP + +S TP+ +P+ + R R R
Sbjct: 28 SSSSSSSSPPPPRRRSSPSP*PPTPAP*SSSSNSDSHTPATSPS-------PSRRSRWRP 186
Query: 104 GTPRPNGETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKV 163
P PN P +SARS S + S S + + + T S ++ R +
Sbjct: 187 PPPPPNPTPPA*GSS--SSARSRSSKSRLRSSKTLPSASSIHTTSPTSSPSATSRRRHRR 360
Query: 164 PATVTTPTIGRGRVNGNSDQTENSISRSGDQHRWPA 199
+T TP+ + S + + I+ WP+
Sbjct: 361 RSTAPTPS-----ASPTSSASSSPIALRTPPSPWPS 453
>TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-like
protein, partial (41%)
Length = 522
Score = 36.6 bits (83), Expect = 0.011
Identities = 40/153 (26%), Positives = 59/153 (38%), Gaps = 1/153 (0%)
Frame = +2
Query: 6 STTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRR-T 64
S T +T +PT T P PS + ++ +R P SSS + +SP T
Sbjct: 101 SNPNKTTSTTSSPTSTTPKPSPSPAPSSTLRAP-------------SSSPASASPASSPT 241
Query: 65 NSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSAR 124
SP + S STP +TP+ S T +P P P + L +R
Sbjct: 242 KSPKPSSPSASAPPSSTP-LTPSTATSESSPTATSSSSSASPAP----PTSFSAL---SR 397
Query: 125 SLSVSFQGESFSIPVSKAKPPSATVTQSLRRST 157
+L + S S P S P + + S S+
Sbjct: 398 ALELKALASSPSPPSSPPPSPPSAICMSTSLSS 496
Score = 30.8 bits (68), Expect = 0.62
Identities = 25/125 (20%), Positives = 46/125 (36%)
Frame = +2
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
T+ +++ T+ K +P+P L + S + P + S S S+S+ SS P
Sbjct: 119 TSTTSSPTSTTPKPSPSPAPSSTLRAPSSSPASASPASSPTKSPKPSSPSASAPPSSTPL 298
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTS 122
++ + +T S+ S P + +P P P +
Sbjct: 299 TPSTATSESSPTATSSSSSASPAPPTSFSALSRALELKALASSPSPPSSPPPSPPSAICM 478
Query: 123 ARSLS 127
+ SLS
Sbjct: 479 STSLS 493
Score = 27.7 bits (60), Expect = 5.3
Identities = 23/77 (29%), Positives = 34/77 (43%), Gaps = 3/77 (3%)
Frame = +2
Query: 309 TNPSSRNGGIGNRSTVP---PKLLVPKKSVFDSPASSPRGIVNNRLQGSPIRSAVRPASP 365
T P S+ N++T P PK S +P+S+ R ++ SP +SP
Sbjct: 77 TKPPSQISSNPNKTTSTTSSPTSTTPKPSPSPAPSSTLRAPSSSPASASPA------SSP 238
Query: 366 SKLGTPSPRSPSRGVSP 382
+K +P P SPS P
Sbjct: 239 TK--SPKPSSPSASAPP 283
>TC18617 weakly similar to UP|Q9FPQ5 (Q9FPQ5) Gamete-specific
hydroxyproline-rich glycoprotein a2, partial (13%)
Length = 578
Score = 36.2 bits (82), Expect = 0.015
Identities = 47/155 (30%), Positives = 62/155 (39%), Gaps = 11/155 (7%)
Frame = +3
Query: 17 APTPTRPPLL-------PSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLV 69
+P+PT PP L PS S + S + S+ + SS SPP T P
Sbjct: 132 SPSPTSPPYLISSSPPSPSASSSPPPGPTPPPPAPSSHSLSTLAGSS-KSPPCLT-PPRH 305
Query: 70 NRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQG---TPRPNGETPVAQKMLFTS-ARS 125
+R QK T S + AA R +S +G P P+G + A + TS +RS
Sbjct: 306 HRHQTPPPQKPTSSASTAATASRPRSASPTGSNRGYPLIPSPHGASSPAPRTCTTSGSRS 485
Query: 126 LSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPER 160
+AKPPS T RRS P R
Sbjct: 486 --------------PRAKPPSKTPP---RRSEPCR 539
>TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, partial (22%)
Length = 505
Score = 35.0 bits (79), Expect = 0.033
Identities = 24/81 (29%), Positives = 36/81 (43%), Gaps = 3/81 (3%)
Frame = +3
Query: 10 TTKNTKRAPTPTRPPLLPSESDNAI---VRKPKAREVTSRYMYSSSSSSSFSSPPRRTNS 66
TT + +P P LPS S A+ P ++ SS++ S S PP +
Sbjct: 144 TTSFSTSSPASHPLPSLPSPSSAAVGPTSSTPPTSPTSAAAATSSATPPSPSPPPTSASP 323
Query: 67 PLVNRTVNSTKQKSTPSVTPA 87
P + T N T S+P+ TP+
Sbjct: 324 PPPSSTPNGTPPSSSPATTPS 386
>TC15474 UP|Q7Y1B9 (Q7Y1B9) Ammonium transporter, complete
Length = 1950
Score = 34.3 bits (77), Expect = 0.056
Identities = 38/136 (27%), Positives = 54/136 (38%), Gaps = 2/136 (1%)
Frame = +3
Query: 4 AISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRR 63
A TT++ + P PT S + N +R P SSS+S SP
Sbjct: 339 AYPTTSSASRSPSEPPPTASSAATSSASNTTLRPPTTTA-------SSSTSGPSPSPLPE 497
Query: 64 TNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTS- 122
+ + R NS+ STP PA ST R G PR G P A + S
Sbjct: 498 SPAAPSPRGHNSSPTSSTPLS*PAL------STRSSRTGSG-PRTAGPAPPAPLAAYYSD 656
Query: 123 -ARSLSVSFQGESFSI 137
AR +S+ + ++S+
Sbjct: 657 PARLISLDPE*STWSV 704
>TC15784 similar to UP|PE31_ARATH (Q9LHA7) Peroxidase 31 precursor (Atperox
P31) (ATP41) , partial (38%)
Length = 475
Score = 33.9 bits (76), Expect = 0.074
Identities = 38/128 (29%), Positives = 50/128 (38%), Gaps = 11/128 (8%)
Frame = +1
Query: 50 SSSSSSSFSSPPRRT----NSPLVNRTVNSTKQKSTPSVT-------PAAFVKRSQSTER 98
SSSS S SSPP T ++ R NS K +TPS T PA S ST
Sbjct: 58 SSSSQHSHSSPPSHTRDSHSTSTNTRAHNSPKSSATPSPTNKSPPPPPAPPRSASSSTTA 237
Query: 99 QRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTP 158
TP + + K +A S S + S S+ KPPS ++ + +
Sbjct: 238 STPTAATPPSSSPQLPSTK---PNATPTSTSPSPATPSTSSSELKPPSNSLAPTPSPAPT 408
Query: 159 ERRKVPAT 166
PAT
Sbjct: 409 SSPPPPAT 432
>TC11455 similar to UP|Q9ZUS8 (Q9ZUS8) At2g37380 protein, partial (10%)
Length = 575
Score = 33.9 bits (76), Expect = 0.074
Identities = 32/145 (22%), Positives = 51/145 (35%)
Frame = +2
Query: 14 TKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVNRTV 73
T +P+P+ LP S + KA S ++ +S S+ SSPP + P
Sbjct: 155 TSNSPSPSHLANLPPHSALPMNSSTKASSYLSTFLPASPWSAPSSSPPPPPHPP------ 316
Query: 74 NSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQGE 133
P TP A S +T P P FTS + + +
Sbjct: 317 --PHPPPPPLETPLAAAHPSTATSPSSPTATPPAPAPSQRTTSSSDFTSTTATTTTHYSN 490
Query: 134 SFSIPVSKAKPPSATVTQSLRRSTP 158
S ++ P + S R+++P
Sbjct: 491 PTSRKLTSTSPSPDSPLFSARKTSP 565
>TC12785 similar to UP|Q7X9B2 (Q7X9B2) 9/13 hydroperoxide lyase, partial
(24%)
Length = 424
Score = 33.9 bits (76), Expect = 0.074
Identities = 33/118 (27%), Positives = 48/118 (39%), Gaps = 4/118 (3%)
Frame = +1
Query: 51 SSSSSSFSSPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNG 110
S SSS + T SP R+ +T +T +VTP++ + ++T + P P+
Sbjct: 64 SRSSSH*NQSQAATASPSSVRSATATTTSTTKAVTPSSPPE*KKTTPLSSEPTCLPAPSY 243
Query: 111 ETPVAQKMLFTSARSLSVSFQGESFSIPVSKA----KPPSATVTQSLRRSTPERRKVP 164
+ T+ RS S S S S S A PPS T S TP+ P
Sbjct: 244 PPTLELLRSSTAPRSPSCSTTTRSRSATSSTALSCPPPPSPAATVSAPT*TPQNLNTP 417
Score = 30.0 bits (66), Expect = 1.1
Identities = 30/106 (28%), Positives = 40/106 (37%), Gaps = 4/106 (3%)
Frame = +1
Query: 6 STTTTTKNTKRAPTPTRPP----LLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPP 61
S T TT +T +A TP+ PP P S+ + P S++ S S
Sbjct: 127 SATATTTSTTKAVTPSSPPE*KKTTPLSSEPTCLPAPSYPPTLELLRSSTAPRSPSCSTT 306
Query: 62 RRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPR 107
R+ S +ST P +PAA V T Q TPR
Sbjct: 307 TRSRS-----ATSSTALSCPPPPSPAATVSAPT*TP---QNLNTPR 420
>TC14277 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (AT1G65720/F1E22_13), partial
(33%)
Length = 941
Score = 33.9 bits (76), Expect = 0.074
Identities = 44/179 (24%), Positives = 64/179 (35%), Gaps = 19/179 (10%)
Frame = +3
Query: 13 NTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVNRT 72
++ +P+P PP P+ + ++ P T S S PR SPL T
Sbjct: 105 SSSSSPSPPPPPHAPAAASSS----PPTPSATLPPTPSPPS-------PRSDPSPLSTST 251
Query: 73 VNSTKQKSTPSVTPAAFVKRSQSTERQRQRQG-----------------TPRPNGETPVA 115
N T ST TP +S + PR + + +
Sbjct: 252 TNPTTSSST---TPKTTTNKSNPNTARLSHAAP*GSPPLLTTTSPPSVTAPRTSSASRLP 422
Query: 116 QKMLFTSARS--LSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTI 172
+A S L + G S S PV+ PP ++ RRST RR PAT +P +
Sbjct: 423 SSSALDAALSPPLPCTSSGPS-SPPVTTTAPPCTAISPMTRRSTAPRR--PATSRSPPL 590
>TC16098 homologue to UP|Q8LAQ5 (Q8LAQ5) Flower pigmentation protein ATAN11,
partial (79%)
Length = 919
Score = 33.1 bits (74), Expect = 0.13
Identities = 44/158 (27%), Positives = 58/158 (35%), Gaps = 5/158 (3%)
Frame = +1
Query: 21 TRPPLLPSESDNAIVRKPKAREVTSRYMYSSSS---SSSFSSPPRRTNSPLVNRTVNSTK 77
TRP P+ S NA P TS +S+ S++ SPP N+P T
Sbjct: 76 TRPKTAPTSSKNAPRSTPTKPHGTSTP*TGASAATKSTASPSPPSSNNTP--------TA 231
Query: 78 QKSTPSVTPAAFVKRSQSTERQRQRQGT--PRPNGETPVAQKMLFTSARSLSVSFQGESF 135
+S+ S TP A RS T T PRP + L +S + S G S
Sbjct: 232 SRSSSSTTPTA---RSDPTRISPSNTHTHPPRPFSSLTKSVNALTSSPPPATSSACGGS- 399
Query: 136 SIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIG 173
P+ PP + S T R P+IG
Sbjct: 400 PTPMKPNPPPPPPASNSRASLTETRTASIVAR*PPSIG 513
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.127 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,932,320
Number of Sequences: 28460
Number of extensions: 132036
Number of successful extensions: 1449
Number of sequences better than 10.0: 179
Number of HSP's better than 10.0 without gapping: 1245
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1377
length of query: 573
length of database: 4,897,600
effective HSP length: 95
effective length of query: 478
effective length of database: 2,193,900
effective search space: 1048684200
effective search space used: 1048684200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC140671.7


