

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
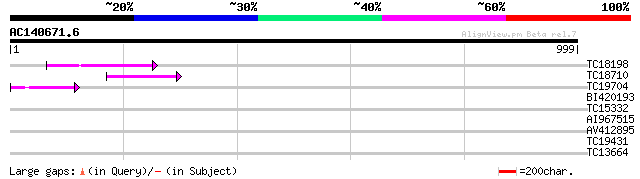
Query= AC140671.6 + phase: 0 /pseudo
(999 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC18198 weakly similar to UP|Q9XGZ5 (Q9XGZ5) T1N24.20 protein, p... 119 2e-27
TC18710 96 2e-20
TC19704 weakly similar to UP|Q9SZ87 (Q9SZ87) RNA-directed DNA po... 76 2e-14
BI420193 32 0.65
TC15332 similar to UP|PPA1_LYCES (P27061) Acid phosphatase precu... 30 1.9
AI967515 29 4.2
AV412895 29 4.2
TC19431 weakly similar to UP|AAR20721 (AAR20721) At5g43820, part... 28 7.2
TC13664 weakly similar to UP|AAR20721 (AAR20721) At5g43820, part... 28 7.2
>TC18198 weakly similar to UP|Q9XGZ5 (Q9XGZ5) T1N24.20 protein, partial
(14%)
Length = 592
Score = 119 bits (299), Expect = 2e-27
Identities = 68/196 (34%), Positives = 110/196 (55%), Gaps = 1/196 (0%)
Frame = -3
Query: 65 RSKWIADGDRNTKYYHSKTIIRRRKNKILTLCDDSSDWIDDPNILKNLVRNFYTDLFKED 124
R K + GD NT+++H+ TI RR N+IL + D W++ + N+++D++
Sbjct: 590 RIK*LK*GDHNTRFFHASTIQRRDFNRILKIKDAHGIWVEGQKKVNEAAVNYFSDIYSS- 414
Query: 125 TPSRDSIVSWTTYPNVV-GKYHNKLISNVQLNECKRALFDMGPHKASGEDGYPAIFFQHC 183
+P + P +V + ++KL + V +E K A+F MG KA G DG +F+Q
Sbjct: 413 SPLQYLRECLAPIPKLVTDRLNHKLCAQVSTDEIKEAVFSMGDLKAPGMDGINGLFYQQN 234
Query: 184 WDKVADSLFNFVKQVWVNHSLISFINNILIVMIPKIEKPEFVSQFRPISLCNVIYKIISK 243
+ V + + N + + + L +N L+ +IPKI E + QFRPIS C+ IYK+ISK
Sbjct: 233 REIVKNDVNNAILDFFDHGELPVELNETLVTLIPKIPHAEAIQQFRPISCCSFIYKVISK 54
Query: 244 VIVNRIKPLLDRIISP 259
+IV R+KP + +ISP
Sbjct: 53 IIVARLKPDMVNLISP 6
>TC18710
Length = 843
Score = 96.3 bits (238), Expect = 2e-20
Identities = 51/134 (38%), Positives = 79/134 (58%), Gaps = 1/134 (0%)
Frame = +2
Query: 171 GEDGYPAIFFQHCWDKVADSLFNFVKQVWVNHSLISFINNILIVMIPKIEKPEFVSQFRP 230
G DG +F+Q D V DS+ + + + IN L+ +IPK+ PE ++QFRP
Sbjct: 188 GMDGMNGLFYQKNGDVVKDSVT*AILAFLERAEIPNEINETLVTLIPKVPHPESINQFRP 367
Query: 231 ISLCNVIYKIISKVIVNRIKPLLDRIISPYQSSFIPGRNIHHNIIVAQEMVHAMSKMKG- 289
IS C +YK+ISK+ V R+K + +ISP QS FI GR I N+++ QE HA+++
Sbjct: 368 ISGCTFLYKVISKIFVARLKDAMPDLISPMQSGFIQGRQIQDNLLIVQEAFHAINRPGAL 547
Query: 290 KKDFMSIKIDL*KS 303
++ IK+D+ K+
Sbjct: 548 GRNHSIIKLDMNKA 589
>TC19704 weakly similar to UP|Q9SZ87 (Q9SZ87) RNA-directed DNA
polymerase-like protein, partial (3%)
Length = 530
Score = 76.3 bits (186), Expect = 2e-14
Identities = 42/124 (33%), Positives = 66/124 (52%), Gaps = 2/124 (1%)
Frame = -3
Query: 1 LKEWNTKTFGNIFRRKNELLARLNGIQNSHNYGYSNFLENLE--KELHEQLAITLYQEEC 58
L EW+ + F + EL +L +QN YSN E+ + K+ +L QEE
Sbjct: 426 LTEWHHQNFSRADKEIAELKNQLEHLQN-----YSNSGEDWQEIKQRKMRLKDLWRQEEL 262
Query: 59 LWFQKSRSKWIADGDRNTKYYHSKTIIRRRKNKILTLCDDSSDWIDDPNILKNLVRNFYT 118
W Q+SR KW+ GD+NTK++H+ T+ RR +N+I + D W+ + V +FY
Sbjct: 261 FWSQRSRIKWLKSGDKNTKFFHASTVQRRERNRIERIKDMQGTWVRSQLAINRDVPDFYP 82
Query: 119 DLFK 122
D++K
Sbjct: 81 DIYK 70
>BI420193
Length = 506
Score = 31.6 bits (70), Expect = 0.65
Identities = 12/30 (40%), Positives = 21/30 (70%)
Frame = -3
Query: 243 KVIVNRIKPLLDRIISPYQSSFIPGRNIHH 272
K+++N I PL+++ S + +F+PG N HH
Sbjct: 375 KLVINNISPLINQCYSS-KGTFVPGTNAHH 289
>TC15332 similar to UP|PPA1_LYCES (P27061) Acid phosphatase precursor 1 ,
partial (89%)
Length = 1040
Score = 30.0 bits (66), Expect = 1.9
Identities = 15/46 (32%), Positives = 27/46 (58%)
Frame = -2
Query: 217 PKIEKPEFVSQFRPISLCNVIYKIISKVIVNRIKPLLDRIISPYQS 262
P I+ P V +P+S+C++I KI K ++N P + II+ + +
Sbjct: 529 PTIKLP-MVKNLQPVSMCSIIGKIREKRLINVENPRVFSIITKFNT 395
>AI967515
Length = 376
Score = 28.9 bits (63), Expect = 4.2
Identities = 15/40 (37%), Positives = 20/40 (49%)
Frame = +2
Query: 63 KSRSKWIADGDRNTKYYHSKTIIRRRKNKILTLCDDSSDW 102
+ S+WI +GDRN + RRR+ K T S DW
Sbjct: 11 RGSSQWIGNGDRNPE----ADAPRRRRTKPSTAASVSGDW 118
>AV412895
Length = 405
Score = 28.9 bits (63), Expect = 4.2
Identities = 23/63 (36%), Positives = 30/63 (47%), Gaps = 3/63 (4%)
Frame = +2
Query: 89 KNKILTLCDDSSDWIDDPNILKNLVRNFYTDLFKEDT---PSRDSIVSWTTYPNVVGKYH 145
KN+IL L S W P I K+L FY D+FK T R S + WT + G
Sbjct: 56 KNQILFLI--RSGWCRLP*I*KHLFGEFYWDVFKLATIC*SGR*STMPWTQSVHYAGWQR 229
Query: 146 NKL 148
N++
Sbjct: 230 NQV 238
>TC19431 weakly similar to UP|AAR20721 (AAR20721) At5g43820, partial (39%)
Length = 603
Score = 28.1 bits (61), Expect = 7.2
Identities = 10/29 (34%), Positives = 19/29 (65%)
Frame = -3
Query: 36 NFLENLEKELHEQLAITLYQEECLWFQKS 64
N L N + ++H+ + L+Q+ CLW Q++
Sbjct: 475 NPLNNNDNKIHKLSRLILHQQSCLWDQRA 389
>TC13664 weakly similar to UP|AAR20721 (AAR20721) At5g43820, partial (17%)
Length = 472
Score = 28.1 bits (61), Expect = 7.2
Identities = 10/29 (34%), Positives = 19/29 (65%)
Frame = -3
Query: 36 NFLENLEKELHEQLAITLYQEECLWFQKS 64
N L N + ++H+ + L+Q+ CLW Q++
Sbjct: 305 NPLNNNDNKIHKLSRLILHQQSCLWDQRA 219
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.360 0.161 0.604
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,488,940
Number of Sequences: 28460
Number of extensions: 297172
Number of successful extensions: 3073
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 2222
Number of HSP's successfully gapped in prelim test: 74
Number of HSP's that attempted gapping in prelim test: 760
Number of HSP's gapped (non-prelim): 2398
length of query: 999
length of database: 4,897,600
effective HSP length: 99
effective length of query: 900
effective length of database: 2,080,060
effective search space: 1872054000
effective search space used: 1872054000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC140671.6


