

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140550.12 + phase: 0
(882 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
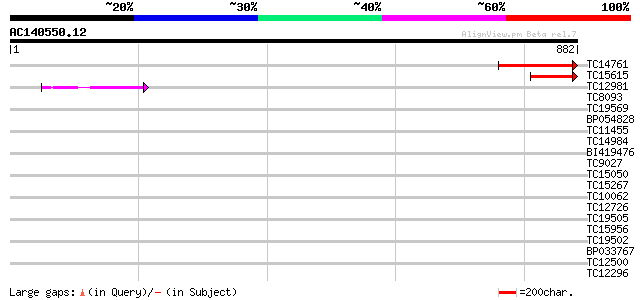
Score E
Sequences producing significant alignments: (bits) Value
TC14761 similar to UP|AAS07369 (AAS07369) Expressed protein, par... 168 3e-42
TC15615 homologue to UP|AAS49490 (AAS49490) DMI1 protein, partia... 136 2e-32
TC12981 44 1e-04
TC8093 homologue to UP|EFTU_TOBAC (P41342) Elongation factor Tu,... 37 0.010
TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor k... 37 0.014
BP054828 37 0.018
TC11455 similar to UP|Q9ZUS8 (Q9ZUS8) At2g37380 protein, partial... 37 0.018
TC14984 similar to GB|AAM62447.1|21553354|AY085214 glycine-rich ... 35 0.052
BI419476 35 0.068
TC9027 weakly similar to UP|AAQ82447 (AAQ82447) SufB, partial (51%) 35 0.068
TC15050 similar to UP|ATP4_PEA (Q41000) ATP synthase delta' chai... 34 0.089
TC15267 similar to UP|EXTN_TOBAC (P13983) Extensin precursor (Ce... 34 0.089
TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein... 34 0.089
TC12726 similar to UP|BCCP_SOYBN (Q42783) Biotin carboxyl carrie... 34 0.089
TC19505 weakly similar to UP|Q9FG35 (Q9FG35) Emb|CAB82953.1, par... 34 0.12
TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid UDP-glu... 33 0.15
TC19502 homologue to PIR|T06435|T06435 ribonuclease S5 homolog -... 33 0.15
BP033767 33 0.15
TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein... 33 0.15
TC12296 weakly similar to UP|Q8S9Y8 (Q8S9Y8) OJ1656_A11.22 prote... 33 0.20
>TC14761 similar to UP|AAS07369 (AAS07369) Expressed protein, partial (14%)
Length = 628
Score = 168 bits (426), Expect = 3e-42
Identities = 86/122 (70%), Positives = 102/122 (83%)
Frame = +3
Query: 761 EILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAE 820
EILD T+NL+S+S+ISDYVLSNELVSMALAMVAED+QIN VLEELFAEEGNEM I+ A+
Sbjct: 3 EILDPXTKNLLSMSKISDYVLSNELVSMALAMVAEDRQINDVLEELFAEEGNEMHIRQAD 182
Query: 821 FYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVFVVLAS 880
YL + EE+ FY+IM+R R R+EI+IGYRLAN ERA+INP K+ RKWSL DVFVV+
Sbjct: 183 IYLREGEEMSFYEIMLRARQRREILIGYRLANAERAVINPPAKTGRRKWSLKDVFVVITE 362
Query: 881 GE 882
E
Sbjct: 363 KE 368
>TC15615 homologue to UP|AAS49490 (AAS49490) DMI1 protein, partial (8%)
Length = 730
Score = 136 bits (342), Expect = 2e-32
Identities = 64/73 (87%), Positives = 69/73 (93%)
Frame = +2
Query: 810 EGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAIINPSEKSVPRKW 869
+GNEMCIKPAEFYLFDQEELCFYDIMIRGR R+EI+IGYRLANQERAIINPSEK V RKW
Sbjct: 74 QGNEMCIKPAEFYLFDQEELCFYDIMIRGRARQEIIIGYRLANQERAIINPSEKLVARKW 253
Query: 870 SLDDVFVVLASGE 882
SL DVFVV+ASG+
Sbjct: 254 SLGDVFVVIASGD 292
>TC12981
Length = 614
Score = 43.9 bits (102), Expect = 1e-04
Identities = 36/173 (20%), Positives = 77/173 (43%), Gaps = 7/173 (4%)
Frame = -2
Query: 50 TTKTDFSEQQWNYPSFLGIGSTSRKRRQPPPPPS---KPPVNLIPPHPRPLSVNDHNKTT 106
T S YPS GI R + P S KP ++++ P +S N+ N +
Sbjct: 463 TNSNTHSAPSSTYPS--GIRHRRRVKFSRTPTTSSNEKPQISIVSDKPSAISKNNLNWLS 290
Query: 107 SSLLPQPSSSSITKQQQQHSTSSPIFYLLVICCIILVPYSAYLQYKLAKLKD--MKLQLC 164
Q + L ++ ++L+ + +L+ ++ KL+ ++L C
Sbjct: 289 -----------------QFGLQFALVTLTIVFLLLLLLRNTHLESQVNKLQGEILRLHAC 161
Query: 165 GQIDFCSRNGKTSIQEEVDDDDNADS--RTIALYIVLFTLILPFVLYKYLDYL 215
Q+D + + T+ + + + ++ R +AL++ L++P +++KY+DY+
Sbjct: 160 HQLDTLNVSSSTAHKSQDTHPCSCENFKRNLALFLSFMLLLIPLIIFKYIDYV 2
>TC8093 homologue to UP|EFTU_TOBAC (P41342) Elongation factor Tu,
chloroplast precursor (EF-Tu), partial (58%)
Length = 1146
Score = 37.4 bits (85), Expect = 0.010
Identities = 31/104 (29%), Positives = 46/104 (43%), Gaps = 1/104 (0%)
Frame = +2
Query: 17 PPLKKTKTLPSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQWNYPSFLGIGSTSRKRR 76
PP T TL L L ++ PP S++ ++ PS L S++R+R
Sbjct: 122 PPNSSTHTLLLLPLTLTPLPP----------ASSSDPNYPPPTSPPPSILPQSSSTRRRN 271
Query: 77 QPPPPPSKPPVNLIPPHP-RPLSVNDHNKTTSSLLPQPSSSSIT 119
PPPP + P PP P PL+ + + S PS++S T
Sbjct: 272 NPPPPSTAAP----PPSPSAPLAASSSARNPMS-TSAPSATSTT 388
Score = 29.6 bits (65), Expect = 2.2
Identities = 29/106 (27%), Positives = 39/106 (36%), Gaps = 3/106 (2%)
Frame = +2
Query: 7 ESSNLNVMNKPPLKKTKT---LPSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQWNYP 63
+SS+ N PP T PS L S + NP + TSTT S +P
Sbjct: 245 QSSSTRRRNNPPPPSTAAPPPSPSAPLAASSSARNPMSTSAPSATSTTAKPPSPPPSPWP 424
Query: 64 SFLGIGSTSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSL 109
S L + + PP K + + P P P S T +L
Sbjct: 425 S-LPSETAPPRNTTKSTPPLKSALAVSPSTPPPSSTRPRIATMLTL 559
>TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor kinase 1;1,
partial (16%)
Length = 486
Score = 37.0 bits (84), Expect = 0.014
Identities = 28/85 (32%), Positives = 37/85 (42%), Gaps = 20/85 (23%)
Frame = +1
Query: 66 LGIGSTSRKRRQPPPPPSK--------PPVNLIPPHPRPLSVNDHNKT---------TSS 108
L S+S PPPPPS+ P + P P P V ++ T TSS
Sbjct: 37 LHTSSSSSSSYSPPPPPSQQPTHSPSTSPTSTTTPPPSPTKVTENPPTAPSTSTKSPTSS 216
Query: 109 LLPQPS---SSSITKQQQQHSTSSP 130
+PS S+ T QQ +HS +SP
Sbjct: 217 ASEEPSPPNPSTSTTQQPKHSPTSP 291
Score = 36.2 bits (82), Expect = 0.023
Identities = 36/122 (29%), Positives = 52/122 (42%), Gaps = 6/122 (4%)
Frame = +1
Query: 15 NKPPLKKTKTL-PSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQWNYPSFLGIGSTSR 73
N P T T P+ + +PPNP+ TSTT QQ + S S
Sbjct: 169 NPPTAPSTSTKSPTSSASEEPSPPNPS-------TSTT------QQPKHSPTSPPASPS- 306
Query: 74 KRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKT-----TSSLLPQPSSSSITKQQQQHSTS 128
P PP +KPP + P PLS + T ++S P P+++S+ K S+S
Sbjct: 307 ----PSPPSTKPPTAMASPSTSPLSATKSHPTPPAAPSASSTPPPTTTSL-KTMSSPSSS 471
Query: 129 SP 130
+P
Sbjct: 472 TP 477
>BP054828
Length = 476
Score = 36.6 bits (83), Expect = 0.018
Identities = 23/51 (45%), Positives = 26/51 (50%), Gaps = 2/51 (3%)
Frame = +2
Query: 79 PPPPSKPPVNLIPPHPRPLSVNDHNKTTSSL--LPQPSSSSITKQQQQHST 127
P PPS+ P P P S N K +S + LPQPSSSS T Q H T
Sbjct: 89 PNPPSRSPT------PNPTSENPKGKLSSPITNLPQPSSSSGTVQPPYHKT 223
>TC11455 similar to UP|Q9ZUS8 (Q9ZUS8) At2g37380 protein, partial (10%)
Length = 575
Score = 36.6 bits (83), Expect = 0.018
Identities = 34/120 (28%), Positives = 48/120 (39%), Gaps = 1/120 (0%)
Frame = +2
Query: 14 MNKPPLKKTKTLPSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQWNYPS-FLGIGSTS 72
+ P L L L+L S P+P+ + S + S + +Y S FL S
Sbjct: 101 LQNPTLSLQHPLTLLHLPTS-NSPSPSHLANLPPHSALPMNSSTKASSYLSTFLPASPWS 277
Query: 73 RKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITKQQQQHSTSSPIF 132
PPPPP PP PP PL+ + TS P+++ Q +TSS F
Sbjct: 278 APSSSPPPPPHPPPHPPPPPLETPLAAAHPSTATSP--SSPTATPPAPAPSQRTTSSSDF 451
>TC14984 similar to GB|AAM62447.1|21553354|AY085214 glycine-rich RNA binding
protein 7 {Arabidopsis thaliana;} , partial (19%)
Length = 483
Score = 35.0 bits (79), Expect = 0.052
Identities = 23/72 (31%), Positives = 31/72 (42%), Gaps = 5/72 (6%)
Frame = +2
Query: 64 SFLGIGSTSRKRRQPPPPP-----SKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSI 118
S + TS R P P S PP PP P PL + + ++L PSS++
Sbjct: 53 SATAVNPTSASRSSPTTAPASLSLSPPPQPSNPPPPPPLPPPEMPSSPATLKSSPSSATA 232
Query: 119 TKQQQQHSTSSP 130
T ST+SP
Sbjct: 233 TAAPSTKSTTSP 268
>BI419476
Length = 520
Score = 34.7 bits (78), Expect = 0.068
Identities = 21/52 (40%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Frame = +1
Query: 70 STSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTT--SSLLPQPSSSSIT 119
S S RR+P PPP P HP P S N +T SSL P P++ S++
Sbjct: 190 SPSPTRRRPSPPP--------PSHPPPTSPASRNGSTRVSSLSPSPTACSVS 321
>TC9027 weakly similar to UP|AAQ82447 (AAQ82447) SufB, partial (51%)
Length = 1016
Score = 34.7 bits (78), Expect = 0.068
Identities = 18/34 (52%), Positives = 22/34 (63%), Gaps = 2/34 (5%)
Frame = +3
Query: 63 PSFLGIGSTSRKRRQPP--PPPSKPPVNLIPPHP 94
P L + S++ R QPP PPP KPP NL+PP P
Sbjct: 66 PRRLPLTSSTHSRPQPPQTPPPPKPP-NLLPPPP 164
>TC15050 similar to UP|ATP4_PEA (Q41000) ATP synthase delta' chain,
mitochondrial precursor , partial (93%)
Length = 951
Score = 34.3 bits (77), Expect = 0.089
Identities = 31/110 (28%), Positives = 50/110 (45%), Gaps = 1/110 (0%)
Frame = +1
Query: 22 TKTLPSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQWNYPSFLGIGSTSRKRRQPPPP 81
+K +P L+LR++ P+D T + S Q ++PS+L PP P
Sbjct: 13 SKLIPHLSLRLA----RPSD-------LTLPSPCSAAQ-HHPSWLA---------PPPTP 129
Query: 82 PSKPPVNLIPPHPRPLS-VNDHNKTTSSLLPQPSSSSITKQQQQHSTSSP 130
PP+ ++PP P P S + +S P+P SSS ++ + S P
Sbjct: 130 ADSPPMWVLPPPPAPTSRMRGRKSALTSTHPRPLSSS*SRALLLRTLSLP 279
>TC15267 similar to UP|EXTN_TOBAC (P13983) Extensin precursor (Cell wall
hydroxyproline-rich glycoprotein), partial (6%)
Length = 1003
Score = 34.3 bits (77), Expect = 0.089
Identities = 32/122 (26%), Positives = 47/122 (38%), Gaps = 2/122 (1%)
Frame = +1
Query: 2 AKSNEESSNLNVMNKPP--LKKTKTLPSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQ 59
+K+N + N + PP + + SL+ +S +PP+ T + S Q
Sbjct: 94 SKTNNNAINTTTVKLPPP*SPSSPSSSSLSPWLSPSPPSSTTPPPPPSHRTNNNNLSTQ- 270
Query: 60 WNYPSFLGIGSTSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSIT 119
P+ +TS R P PPS PP P H P S + +S P P S
Sbjct: 271 ---PTPSAPSATSLASRTPASPPSPPPTTTPPTH-NPSSPSPSAPPWTSSPPSPPRSESR 438
Query: 120 KQ 121
Q
Sbjct: 439 PQ 444
Score = 28.5 bits (62), Expect = 4.9
Identities = 25/95 (26%), Positives = 37/95 (38%), Gaps = 12/95 (12%)
Frame = +1
Query: 48 TSTTKTDFSEQQWNYPSFLGIGSTSRKRRQPPPPPS------------KPPVNLIPPHPR 95
T+TT T S+ N I +T+ K P P S PP + PP P
Sbjct: 70 TTTTTTTTSKTNNN-----AINTTTVKLPPP*SPSSPSSSSLSPWLSPSPPSSTTPPPPP 234
Query: 96 PLSVNDHNKTTSSLLPQPSSSSITKQQQQHSTSSP 130
N++N +T PS++S+ + S P
Sbjct: 235 SHRTNNNNLSTQPTPSAPSATSLASRTPASPPSPP 339
>TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein 2, partial
(66%)
Length = 481
Score = 34.3 bits (77), Expect = 0.089
Identities = 21/53 (39%), Positives = 25/53 (46%)
Frame = -1
Query: 78 PPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITKQQQQHSTSSP 130
P PPP +PP PP P + TS L +PS S T +STSSP
Sbjct: 322 P*PPPPRPPPPAPPPRRVPCTAWPLGPVTSIALARPSGSEAT----MNSTSSP 176
Score = 31.6 bits (70), Expect = 0.58
Identities = 11/19 (57%), Positives = 13/19 (67%)
Frame = -1
Query: 78 PPPPPSKPPVNLIPPHPRP 96
PPPPP +PP PP+P P
Sbjct: 478 PPPPPPRPPPRPPPPYPPP 422
Score = 28.5 bits (62), Expect = 4.9
Identities = 11/21 (52%), Positives = 13/21 (61%)
Frame = -1
Query: 76 RQPPPPPSKPPVNLIPPHPRP 96
R PPP P PP +PP+P P
Sbjct: 448 RPPPPYPPPPP*PPLPPYPPP 386
Score = 27.7 bits (60), Expect = 8.3
Identities = 11/19 (57%), Positives = 11/19 (57%)
Frame = -1
Query: 78 PPPPPSKPPVNLIPPHPRP 96
PPPPP PP PP P P
Sbjct: 394 PPPPPYPPPPRPPPP*PPP 338
>TC12726 similar to UP|BCCP_SOYBN (Q42783) Biotin carboxyl carrier protein
of acetyl-CoA carboxylase, chloroplast precursor (BCCP)
, partial (8%)
Length = 393
Score = 34.3 bits (77), Expect = 0.089
Identities = 17/36 (47%), Positives = 23/36 (63%), Gaps = 2/36 (5%)
Frame = -2
Query: 79 PPPPSKPPV--NLIPPHPRPLSVNDHNKTTSSLLPQ 112
PPPP+ PP+ +LIPP P LS + N T S+ P+
Sbjct: 296 PPPPNPPPIFASLIPPTPTFLSASLLNPTPSAFSPR 189
>TC19505 weakly similar to UP|Q9FG35 (Q9FG35) Emb|CAB82953.1, partial (12%)
Length = 723
Score = 33.9 bits (76), Expect = 0.12
Identities = 27/105 (25%), Positives = 43/105 (40%), Gaps = 4/105 (3%)
Frame = +1
Query: 62 YPSFLGIGSTSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITKQ 121
+P L + PPPPP PP PP P PL++N P S + +
Sbjct: 388 FPLLLTLHLPGSPTSSPPPPPPPPPP---PPPPVPLNLN-----CLPFSPTFSLTPLLHP 543
Query: 122 QQQHSTSSPIFYLLVICCIILVPYSAYLQYK----LAKLKDMKLQ 162
Q H + + L ++ +L+P A ++ KL+D +Q
Sbjct: 544 QILHRRRATVLDLAMLLLHLLIPKPAQKHWQQPLHKTKLRDQ*IQ 678
>TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid
UDP-glucosyltransferase (Limonoid glucosyltransferase)
(Limonoid GTase) (LGTase) , partial (65%)
Length = 1169
Score = 33.5 bits (75), Expect = 0.15
Identities = 21/54 (38%), Positives = 28/54 (50%), Gaps = 1/54 (1%)
Frame = +3
Query: 65 FLGIGSTSRKRRQPPP-PPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSS 117
F G S S +R P P PP KPP N P P P S H ++++L +SS+
Sbjct: 84 FSG*PSASPQRAPPSPSPPHKPPAN--PCEPPPTSPTKHPLQSATVLSSSNSST 239
>TC19502 homologue to PIR|T06435|T06435 ribonuclease S5 homolog - garden pea
{Pisum sativum;}, partial (5%)
Length = 516
Score = 33.5 bits (75), Expect = 0.15
Identities = 23/97 (23%), Positives = 42/97 (42%), Gaps = 7/97 (7%)
Frame = +3
Query: 47 GTSTTKTDFSEQQWNYPSFLGIGSTSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTT 106
G S W YP +T+R PPP + + PP P+P S + + T
Sbjct: 102 GRSQVPAPTPSTSWRYPQ---PWNTARSA----PPPRRSRRSSPPPFPKPSSTSSTTRAT 260
Query: 107 SSLLPQPSSSSITKQQQQHST-------SSPIFYLLV 136
S++ PS++ +++ ++ + PIF+L +
Sbjct: 261 SAVTALPSAAPSSRRPMWRNS*RRGRHLTFPIFHLFI 371
>BP033767
Length = 541
Score = 33.5 bits (75), Expect = 0.15
Identities = 17/41 (41%), Positives = 23/41 (55%), Gaps = 1/41 (2%)
Frame = +1
Query: 78 PPPPPSKPPVN-LIPPHPRPLSVNDHNKTTSSLLPQPSSSS 117
PP PP+ PP + L PP+P P + + + S P PSS S
Sbjct: 343 PPSPPTSPPSSSLKPPNPNPPTSSSASPLPPSPAPSPSSLS 465
>TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein {Danio
rerio;} , partial (5%)
Length = 678
Score = 33.5 bits (75), Expect = 0.15
Identities = 25/92 (27%), Positives = 35/92 (37%)
Frame = +1
Query: 8 SSNLNVMNKPPLKKTKTLPSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQWNYPSFLG 67
S+ ++ ++ PL T P + PP P+ G +K S
Sbjct: 10 STPMSSADRAPLPPPSTPPKSLKALPPPPPPPSAPFIAGSAIISKVSKS----------- 156
Query: 68 IGSTSRKRRQPPPPPSKPPVNLIPPHPRPLSV 99
+G S PPPP PP +L P P P SV
Sbjct: 157VGDVSLPCSPSPPPPQPPPPSLPPTTPPPASV 252
Score = 33.1 bits (74), Expect = 0.20
Identities = 22/55 (40%), Positives = 26/55 (47%), Gaps = 9/55 (16%)
Frame = +1
Query: 77 QPPPPPSKPPVNLIPPHPRPLSVNDHNKT---------TSSLLPQPSSSSITKQQ 122
QPPPPP K V IPP P PL ++ ++ TS P P SS T Q
Sbjct: 316 QPPPPPPKYGVTTIPPSP-PLLMSATDRAPLPPPSTHLTSLTTPSPPSSRTTITQ 477
Score = 32.7 bits (73), Expect = 0.26
Identities = 19/40 (47%), Positives = 22/40 (54%)
Frame = +1
Query: 77 QPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSS 116
QPP PP K V IPP P P+S D +T LL P +S
Sbjct: 499 QPPSPPPKYGVATIPPPPLPMSSID--RTPLPLLSTPLTS 612
>TC12296 weakly similar to UP|Q8S9Y8 (Q8S9Y8) OJ1656_A11.22 protein, partial
(10%)
Length = 613
Score = 33.1 bits (74), Expect = 0.20
Identities = 23/57 (40%), Positives = 27/57 (47%), Gaps = 3/57 (5%)
Frame = +1
Query: 77 QPPPPPSKPP--VNLIPPHPRPLSVN-DHNKTTSSLLPQPSSSSITKQQQQHSTSSP 130
QPPP P+ PP +L P P P S D + S LLP PS S + H SP
Sbjct: 16 QPPPSPASPPGSPSLAPRRPPPGSPPCDPSTPPSCLLPWPSPSWRWRVAPPHRMRSP 186
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.137 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,721,950
Number of Sequences: 28460
Number of extensions: 241515
Number of successful extensions: 4726
Number of sequences better than 10.0: 257
Number of HSP's better than 10.0 without gapping: 3017
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3955
length of query: 882
length of database: 4,897,600
effective HSP length: 98
effective length of query: 784
effective length of database: 2,108,520
effective search space: 1653079680
effective search space used: 1653079680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC140550.12


