

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140549.6 - phase: 0 /pseudo
(305 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
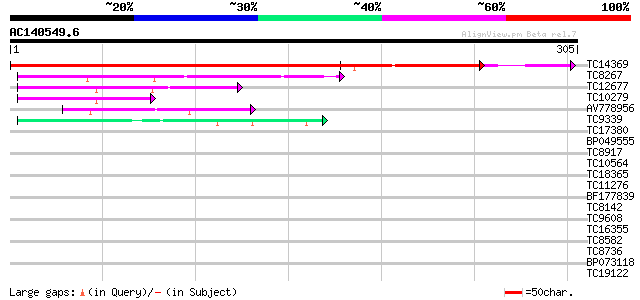
Score E
Sequences producing significant alignments: (bits) Value
TC14369 similar to UP|Q8S3W1 (Q8S3W1) Elongation factor 1-gamma,... 403 e-113
TC8267 similar to UP|Q948X4 (Q948X4) Glutathione S-transferase, ... 62 2e-10
TC12677 similar to UP|Q9FQD6 (Q9FQD6) Glutathione S-transferase ... 60 6e-10
TC10279 similar to UP|Q9FQD6 (Q9FQD6) Glutathione S-transferase ... 50 5e-07
AV778956 46 9e-06
TC9339 similar to UP|Q9FQE3 (Q9FQE3) Glutathione S-transferase G... 40 4e-04
TC17380 similar to UP|Q84UH4 (Q84UH4) Dehydroascorbate reductase... 39 0.001
BP049555 36 0.007
TC8917 similar to UP|AAH64262 (AAH64262) MGC76273 protein, parti... 34 0.027
TC10564 similar to UP|Q9FQD3 (Q9FQD3) Glutathione S-transferase ... 33 0.046
TC18365 similar to UP|Q940G9 (Q940G9) Periaxin-like protein, par... 33 0.046
TC11276 weakly similar to UP|HDA1_CHICK (P56517) Histone deacety... 33 0.046
BF177839 33 0.060
TC8142 UP|BAD11133 (BAD11133) Glutamate-rich protein, complete 33 0.078
TC9608 similar to UP|Q94AZ4 (Q94AZ4) At1g12310/F5O11_2, partial ... 33 0.078
TC16355 similar to UP|Q9M4U6 (Q9M4U6) Histone H1 variant, partia... 32 0.10
TC8582 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetyla... 32 0.13
TC8736 similar to PIR|T06396|T06396 isoprenylated protein - soyb... 32 0.17
BP073118 32 0.17
TC19122 similar to UP|Q9LXI5 (Q9LXI5) Zinc finger-like protein, ... 31 0.30
>TC14369 similar to UP|Q8S3W1 (Q8S3W1) Elongation factor 1-gamma, complete
Length = 1710
Score = 403 bits (1035), Expect = e-113
Identities = 205/258 (79%), Positives = 223/258 (85%), Gaps = 3/258 (1%)
Frame = +1
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNN-LFGSSLIHQAQID 59
MGVSNKTP+F+ MNPIGKVPVLETPDGPVFESNAIARYVAR +N L+GSSLI AQI+
Sbjct: 253 MGVSNKTPEFLKMNPIGKVPVLETPDGPVFESNAIARYVARSKDDNTLYGSSLIEYAQIE 432
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QWIDF+S+EIDANI K Y PR G+A + PP EE AI+SLKRAL ALNSHLA NTYLVGHS
Sbjct: 433 QWIDFASMEIDANIAKWYYPRAGYAPFLPPAEEFAITSLKRALSALNSHLATNTYLVGHS 612
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
VTLADII T NLY+GFA +LVKSFTSEFPHVERYFWTLVNQPNFRKI+GQVKQ EA+P +
Sbjct: 613 VTLADIILTCNLYLGFAYILVKSFTSEFPHVERYFWTLVNQPNFRKIVGQVKQAEAVPAV 792
Query: 180 PSAKQ--QPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDD 237
SAK+ QPKESKPK K EPK VAK EPE PK E EE APKPK KNPLDLLPPSKM+LD+
Sbjct: 793 QSAKKPSQPKESKPKAKSEPKAVAKKEPE-PKEEEEEVAPKPKAKNPLDLLPPSKMVLDE 969
Query: 238 WKRLYSNTKRNFREVAIK 255
WKRLYSNTK NFREVAIK
Sbjct: 970 WKRLYSNTKTNFREVAIK 1023
Score = 46.6 bits (109), Expect = 5e-06
Identities = 38/126 (30%), Positives = 52/126 (41%)
Frame = +2
Query: 179 IPSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDDW 238
+PS + + + P+K + PK + P PKP+ PL + W
Sbjct: 809 LPSQRNLSPRQRVSLRQWPRK-----SQSPKKKKRR*PPSPKPRIPLIFFLQVRWF---W 964
Query: 239 KRLYSNTKRNFREVAIKDCSKECQNLLTRFNDLIPLFDLQDSGTCMIQRDTLSGSVITNT 298
+T +K S Q+ +D GTCMI RDTL GSVITNT
Sbjct: 965 MSGKGSTP------TLKPTSVRSQS--------------KDFGTCMIPRDTLCGSVITNT 1084
Query: 299 MMRTLF 304
+RTLF
Sbjct: 1085TVRTLF 1102
>TC8267 similar to UP|Q948X4 (Q948X4) Glutathione S-transferase, complete
Length = 995
Score = 61.6 bits (148), Expect = 2e-10
Identities = 51/184 (27%), Positives = 77/184 (41%), Gaps = 8/184 (4%)
Frame = +1
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVA---RLGHNNLFGSSLIHQAQIDQW 61
+K F+++NP G+VP LE D +FES AI +Y+A L A W
Sbjct: 238 HKKEPFLSLNPFGQVPALEDGDLKLFESRAITKYIAYEYADKGTELLSKDSKKMAVTTVW 417
Query: 62 IDFSSLEIDANIMKL-----YLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLV 116
++ + D KL + P G T VEEN + L L+ L+ + YL
Sbjct: 418 LEVEAHHFDQPSSKLAFELAFKPMFGMTTDPAVVEENE-AKLAGVLDVYEKRLSGSKYLA 594
Query: 117 GHSVTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAM 176
TLAD+ N++ K F S PHV + + +P + K++ +
Sbjct: 595 AECFTLADLHHIPNIFYLMNTQSKKLFESR-PHVNAWVADITARPAWSKVVAM------L 753
Query: 177 PPIP 180
PP P
Sbjct: 754 PPKP 765
>TC12677 similar to UP|Q9FQD6 (Q9FQD6) Glutathione S-transferase GST 22
(Fragment) , complete
Length = 876
Score = 59.7 bits (143), Expect = 6e-10
Identities = 37/129 (28%), Positives = 64/129 (48%), Gaps = 8/129 (6%)
Frame = +3
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHN---NLFGSSLIHQAQIDQW 61
+K P+++ + P G++PV++ D ++ES AI RY A N L G ++ + ++QW
Sbjct: 144 HKDPEYLKIQPFGELPVIQDGDYTLYESRAIIRYYAEKYKNQGTELLGKTIEERGLVEQW 323
Query: 62 IDFSSLEIDANIMK-----LYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLV 116
+D + I L+ P LG + P V E + L + L+ L+ + YL
Sbjct: 324 LDVEAHSFQPAIHNLVNHVLFAPILGTPS-DPKVIEESDEKLVKVLDIYEERLSKSKYLA 500
Query: 117 GHSVTLADI 125
G +LAD+
Sbjct: 501 GDFFSLADL 527
>TC10279 similar to UP|Q9FQD6 (Q9FQD6) Glutathione S-transferase GST 22
(Fragment) , partial (60%)
Length = 505
Score = 50.1 bits (118), Expect = 5e-07
Identities = 23/77 (29%), Positives = 42/77 (53%), Gaps = 3/77 (3%)
Frame = +1
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHN---NLFGSSLIHQAQIDQW 61
NK P+++ + P GK+PV++ D ++ES AI RY A N L G ++ + ++QW
Sbjct: 223 NKEPEYLKLQPFGKLPVIQDGDYTLYESRAIIRYYAEKYRNQGTELLGKTVEERGLVEQW 402
Query: 62 IDFSSLEIDANIMKLYL 78
++ + ++ L L
Sbjct: 403 LEVEAHNFHPPVLDLVL 453
>AV778956
Length = 545
Score = 45.8 bits (107), Expect = 9e-06
Identities = 32/113 (28%), Positives = 53/113 (46%), Gaps = 9/113 (7%)
Frame = -2
Query: 29 VFESNAIARYVARL----GHNNLFGSSLIHQAQIDQWIDFSSLEIDANIMKLYLPRLGFA 84
+FES AI RY+ G+ L+G++ + +A IDQW++ + L +L FA
Sbjct: 544 LFESRAICRYICERHKEKGNKELYGTNPLAKASIDQWLEAEGQSFNPPSSTLVF-QLAFA 368
Query: 85 TYFP-PVEENAI----SSLKRALEALNSHLAHNTYLVGHSVTLADIITTSNLY 132
P +E AI L + L+ + L +L G +LAD+ N++
Sbjct: 367 PRMKIPQDEGAIKQNQEKLAKVLDVYDKRLGETRFLAGDEFSLADLSHLPNIH 209
>TC9339 similar to UP|Q9FQE3 (Q9FQE3) Glutathione S-transferase GST 15
(Fragment) , partial (44%)
Length = 981
Score = 40.4 bits (93), Expect = 4e-04
Identities = 45/181 (24%), Positives = 73/181 (39%), Gaps = 14/181 (7%)
Frame = +3
Query: 5 NKTPQFINMNPI-GKVPVLETPDGPVFESNAIARYVARL-GHNNLFGSSLIHQAQIDQWI 62
NK+PQ + NP+ K PVL P+ ES I Y+ L N L S +A W+
Sbjct: 120 NKSPQLLQYNPVHKKTPVLVHDGKPLCESMIIVEYIDELWPQNPLVPSDPYEKAVARFWV 299
Query: 63 DFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLA--HNTYLVGHSV 120
+ ++M LP+ F + ++ I + E + H +L G ++
Sbjct: 300 RYVD-----DMMSPVLPQF-FGRFGSEEQDKVIKDIWERFEVIEDQYLGDHKKFLGGDTL 461
Query: 121 TLADIITTS--NLYIGFAKLL-VKSFTSE-FPHVERYFWTLV------NQPNFRKILGQV 170
+ DI S +G L VK +E FP + +F + N P K++ +
Sbjct: 462 NIVDITFGSFMKTLVGMQDALEVKILVAEKFPRLHAWFNNFMDVPVINNNPEHEKLVAAM 641
Query: 171 K 171
K
Sbjct: 642 K 644
>TC17380 similar to UP|Q84UH4 (Q84UH4) Dehydroascorbate reductase, complete
Length = 875
Score = 38.5 bits (88), Expect = 0.001
Identities = 43/167 (25%), Positives = 67/167 (39%), Gaps = 4/167 (2%)
Frame = +2
Query: 3 VSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNNLFGSSLIHQAQIDQWI 62
+SNK F+++NP GKVPV++ D V +S+ I + SL+
Sbjct: 218 LSNKPQWFLDVNPEGKVPVVKFGDKWVADSDVIVGIL----EEKYPEPSLVTPP------ 367
Query: 63 DFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHL-AHNTYLVGHSVT 121
+F+S + + I ++ L + +L L AL+ HL AH Y+ G VT
Sbjct: 368 EFAS--VGSKIFGSFVSFL----KSKDANDGTEQALVAELSALDEHLKAHGPYVAGERVT 529
Query: 122 LADIITTSNLY---IGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRK 165
D+ LY + S HV Y L + +F K
Sbjct: 530 AVDLSLAPKLYHLVVALDHFKKWSIPESLAHVHNYVKLLFARESFEK 670
>BP049555
Length = 555
Score = 36.2 bits (82), Expect = 0.007
Identities = 22/65 (33%), Positives = 35/65 (53%), Gaps = 5/65 (7%)
Frame = +2
Query: 160 QPNFRKILGQVKQTEAMPPIPSAKQQPKE-----SKPKTKDEPKKVAKSEPEKPKVEVEE 214
Q F + +G+ EA P A+ +P+E ++ K +++P K E EKP+ E +E
Sbjct: 203 QSTFLEQMGE----EAKQEQPQAEAKPEEKTEEKAEEKAEEKPAAEEKKEEEKPEEEKKE 370
Query: 215 EAPKP 219
E PKP
Sbjct: 371 EEPKP 385
Score = 34.3 bits (77), Expect = 0.027
Identities = 25/78 (32%), Positives = 39/78 (49%), Gaps = 3/78 (3%)
Frame = +2
Query: 161 PNFRKILGQVKQT--EAMPPIPSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPK 218
P R ++ ++ T E M +Q E+KP+ K E K K+E EKP E ++E K
Sbjct: 173 PKIRIVI*YIQSTFLEQMGEEAKQEQPQAEAKPEEKTEEKAEEKAE-EKPAAEEKKEEEK 349
Query: 219 P-KPKNPLDLLPPSKMIL 235
P + K + PP+ +L
Sbjct: 350 PEEEKKEEEPKPPAPCVL 403
>TC8917 similar to UP|AAH64262 (AAH64262) MGC76273 protein, partial (3%)
Length = 676
Score = 34.3 bits (77), Expect = 0.027
Identities = 18/55 (32%), Positives = 28/55 (50%), Gaps = 8/55 (14%)
Frame = +2
Query: 172 QTEAMPPIPSAKQQPKESKPKTKD--------EPKKVAKSEPEKPKVEVEEEAPK 218
+ EA P + +PKE K T++ PK + + E+PK +V+EE PK
Sbjct: 302 EEEAKKPEEEVQAEPKEEKATTEEVKVETKEVNPKPEEEPKEEEPKAQVQEEKPK 466
Score = 26.9 bits (58), Expect = 4.3
Identities = 13/38 (34%), Positives = 25/38 (65%)
Frame = +2
Query: 183 KQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPK 220
+++ K+ + + + EPK+ K+ E+ KVE +E PKP+
Sbjct: 302 EEEAKKPEEEVQAEPKE-EKATTEEVKVETKEVNPKPE 412
Score = 26.6 bits (57), Expect = 5.6
Identities = 15/52 (28%), Positives = 24/52 (45%)
Frame = +2
Query: 160 QPNFRKILGQVKQTEAMPPIPSAKQQPKESKPKTKDEPKKVAKSEPEKPKVE 211
+P K + + E P +++PKE +PK A+ + EKPK E
Sbjct: 341 EPKEEKATTEEVKVETKEVNPKPEEEPKEEEPK--------AQVQEEKPKTE 472
>TC10564 similar to UP|Q9FQD3 (Q9FQD3) Glutathione S-transferase GST 25 ,
partial (53%)
Length = 500
Score = 33.5 bits (75), Expect = 0.046
Identities = 14/32 (43%), Positives = 21/32 (64%)
Frame = +1
Query: 8 PQFINMNPIGKVPVLETPDGPVFESNAIARYV 39
P+F+ +NP+G VPVL +F+S AI Y+
Sbjct: 271 PEFLKLNPVGCVPVLVDGPAVIFDSFAIIMYL 366
>TC18365 similar to UP|Q940G9 (Q940G9) Periaxin-like protein, partial (19%)
Length = 404
Score = 33.5 bits (75), Expect = 0.046
Identities = 25/75 (33%), Positives = 35/75 (46%), Gaps = 6/75 (8%)
Frame = +3
Query: 156 TLVNQPNFRKILGQVKQTEAMPPIPSAKQQPKESKPKTKDEPKKVAKS-----EPEKPKV 210
TL++ + + G E+ P +P + PK PK + PK +PE PKV
Sbjct: 126 TLLSANSHIMVAGARILLESKPVLPEVPELPKPELPKIPEFPKPELPKLPELPKPELPKV 305
Query: 211 EVEEEAPKPK-PKNP 224
V E PKP+ PK P
Sbjct: 306 NV-PELPKPELPKVP 347
Score = 30.8 bits (68), Expect = 0.30
Identities = 20/53 (37%), Positives = 26/53 (48%), Gaps = 5/53 (9%)
Frame = +3
Query: 177 PPIPSAKQQPKESKPKTKDEPK----KVAKSEPEKPKVEVEEEAPKPK-PKNP 224
P +P + PK PK + PK KV E KP++ E PKP+ PK P
Sbjct: 222 PELPKIPEFPKPELPKLPELPKPELPKVNVPELPKPELPKVPELPKPELPKVP 380
>TC11276 weakly similar to UP|HDA1_CHICK (P56517) Histone deacetylase 1
(HD1), partial (5%)
Length = 636
Score = 33.5 bits (75), Expect = 0.046
Identities = 16/39 (41%), Positives = 25/39 (64%)
Frame = +3
Query: 180 PSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPK 218
P K++PK+ +P+ ++E K KSE E K E ++E PK
Sbjct: 27 PEKKEEPKKEEPQKEEEKKDEPKSE-EPQKEEAKKEEPK 140
Score = 31.6 bits (70), Expect = 0.17
Identities = 14/39 (35%), Positives = 22/39 (55%)
Frame = +3
Query: 183 KQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKP 221
K +PK +P+ ++ K+ K E EK + EEE K +P
Sbjct: 81 KDEPKSEEPQKEEAKKEEPKKEEEKKDEKKEEEKKKEQP 197
>BF177839
Length = 449
Score = 33.1 bits (74), Expect = 0.060
Identities = 28/102 (27%), Positives = 49/102 (47%), Gaps = 1/102 (0%)
Frame = +2
Query: 129 SNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIPSAKQQPKE 188
S++ + A++L++S P + + L+ +P ++ K P +P P+
Sbjct: 158 SHMMVAGARILLESSEPHVPQLPKPELPLLQKPALPEVPELPKPELPKPELPKVNV-PEL 334
Query: 189 SKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPK-PKNPLDLLP 229
KP+ PK ++PE PK+ V E PKP+ PK + LP
Sbjct: 335 PKPEL---PKVPELAKPELPKINV-PELPKPELPKVNVPELP 448
>TC8142 UP|BAD11133 (BAD11133) Glutamate-rich protein, complete
Length = 999
Score = 32.7 bits (73), Expect = 0.078
Identities = 21/60 (35%), Positives = 33/60 (55%), Gaps = 1/60 (1%)
Frame = +1
Query: 169 QVKQTEAMPPIPSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAP-KPKPKNPLDL 227
Q TE P A + P ++P T +EPK+ EP+ + E E EAP P+ ++PL++
Sbjct: 187 QPAATEVAAPEQPAAEVPAPAEPAT-EEPKEETTEEPK--ETETETEAPVAPETQDPLEV 357
Score = 31.2 bits (69), Expect = 0.23
Identities = 16/55 (29%), Positives = 29/55 (52%), Gaps = 7/55 (12%)
Frame = +1
Query: 185 QPKESKPKTKDEPKKVAKSEPEKPKVEVE-------EEAPKPKPKNPLDLLPPSK 232
+P+E KP+T+ +K + + E+P E EE KP + P++++P K
Sbjct: 385 KPEEVKPETEKTEEKTEEPKTEEPAATTETESAAPVEEENKPAEEAPVEVVPVEK 549
Score = 26.2 bits (56), Expect = 7.3
Identities = 21/63 (33%), Positives = 32/63 (50%), Gaps = 16/63 (25%)
Frame = +1
Query: 178 PIPSAKQQPKESK--------PKTKD----EPKKV---AKSEPEKPKVE-VEEEAPKPKP 221
P ++PKE++ P+T+D E K+V AK E KP+ E EE+ +PK
Sbjct: 268 PKEETTEEPKETETETEAPVAPETQDPLEVENKEVTEEAKPEEVKPETEKTEEKTEEPKT 447
Query: 222 KNP 224
+ P
Sbjct: 448 EEP 456
>TC9608 similar to UP|Q94AZ4 (Q94AZ4) At1g12310/F5O11_2, partial (97%)
Length = 817
Score = 32.7 bits (73), Expect = 0.078
Identities = 20/54 (37%), Positives = 24/54 (44%), Gaps = 5/54 (9%)
Frame = +1
Query: 190 KPKTKDEPKKVAKSEP---EKPKVEVEEEAPKPK--PKNPLDLLPPSKMILDDW 238
KP P A+S P E V EE PKP P +P + PP + LD W
Sbjct: 148 KPSHSSTPTATARSPPLSSESSCVPSEETLPKPSSNPSSPRRISPPPSISLDFW 309
>TC16355 similar to UP|Q9M4U6 (Q9M4U6) Histone H1 variant, partial (50%)
Length = 584
Score = 32.3 bits (72), Expect = 0.10
Identities = 22/60 (36%), Positives = 29/60 (47%), Gaps = 1/60 (1%)
Frame = +1
Query: 162 NFRKILG-QVKQTEAMPPIPSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPK 220
NF+KILG Q+K A + K K S K KVAK++ EK + AP+ K
Sbjct: 328 NFKKILGLQLKNQAAKGKLVKIKASYKLSGAKKDGAGAKVAKAKTEKKDAAAAKVAPRQK 507
Score = 25.8 bits (55), Expect = 9.6
Identities = 11/33 (33%), Positives = 20/33 (60%)
Frame = +1
Query: 178 PIPSAKQQPKESKPKTKDEPKKVAKSEPEKPKV 210
P+ K PKE+KP + +PK + +P++ K+
Sbjct: 106 PVEEVKA-PKETKPAKEKKPKAPKEKKPKQGKI 201
>TC8582 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (65%)
Length = 1341
Score = 32.0 bits (71), Expect = 0.13
Identities = 17/47 (36%), Positives = 23/47 (48%), Gaps = 2/47 (4%)
Frame = +1
Query: 179 IPSAKQQPKESKPKTKDEPKKVAKSEPEKP--KVEVEEEAPKPKPKN 223
+ K++P ES KT KK + PEKP K V P PK ++
Sbjct: 682 VDQGKKRPNESASKTPVSGKKAKNATPEKPDGKKNVHTATPHPKKQD 822
>TC8736 similar to PIR|T06396|T06396 isoprenylated protein - soybean
(fragment) {Glycine max;}, partial (84%)
Length = 1268
Score = 31.6 bits (70), Expect = 0.17
Identities = 21/57 (36%), Positives = 32/57 (55%), Gaps = 4/57 (7%)
Frame = +2
Query: 171 KQTEAMPPIPSAKQQPKES-KPKTKDEPKKVAKSEPEKPKV---EVEEEAPKPKPKN 223
K+ EA P + ++ K+ KPK EP+K K + EKPK E +++ K KPK+
Sbjct: 488 KKKEADKPKAAEHEKKKDGEKPKAAAEPEK--KKDSEKPKAAEPEKKKDGDKEKPKS 652
Score = 29.3 bits (64), Expect = 0.87
Identities = 21/64 (32%), Positives = 33/64 (50%), Gaps = 6/64 (9%)
Frame = +2
Query: 168 GQVKQTEAMPPIPSAKQQPKESKP-KTKDEPKKVAKSE-----PEKPKVEVEEEAPKPKP 221
G+ + A P ++PK ++P K KD K+ KS+ EKPK ++ AP P P
Sbjct: 542 GEKPKAAAEPEKKKDSEKPKAAEPEKKKDGDKEKPKSDGPKKGDEKPK---DKPAPGPIP 712
Query: 222 KNPL 225
+P+
Sbjct: 713 VHPV 724
>BP073118
Length = 495
Score = 31.6 bits (70), Expect = 0.17
Identities = 21/62 (33%), Positives = 29/62 (45%), Gaps = 2/62 (3%)
Frame = +1
Query: 5 NKTPQFINMNPI-GKVPVLETPDGPVFESNAIARYVARL-GHNNLFGSSLIHQAQIDQWI 62
NK+PQ + NP+ K PVL P+ ES I Y+ L N L S +A W+
Sbjct: 109 NKSPQLLQYNPVHKKTPVLVHDGKPLCESMIIVEYIDELWPQNPLVPSDPYEKAVARFWV 288
Query: 63 DF 64
+
Sbjct: 289 RY 294
>TC19122 similar to UP|Q9LXI5 (Q9LXI5) Zinc finger-like protein, partial
(58%)
Length = 737
Score = 30.8 bits (68), Expect = 0.30
Identities = 14/41 (34%), Positives = 23/41 (55%), Gaps = 1/41 (2%)
Frame = -3
Query: 172 QTEAMPPIPSAKQQPKESKPKTKDEPKK-VAKSEPEKPKVE 211
+T + PP P+A+++P++ +P PKK A PE E
Sbjct: 489 RTGSAPPSPTAQKEPQQQQPAPLSSPKKSTAAGSPETATTE 367
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,155,755
Number of Sequences: 28460
Number of extensions: 74334
Number of successful extensions: 659
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 609
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 645
length of query: 305
length of database: 4,897,600
effective HSP length: 90
effective length of query: 215
effective length of database: 2,336,200
effective search space: 502283000
effective search space used: 502283000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC140549.6


