

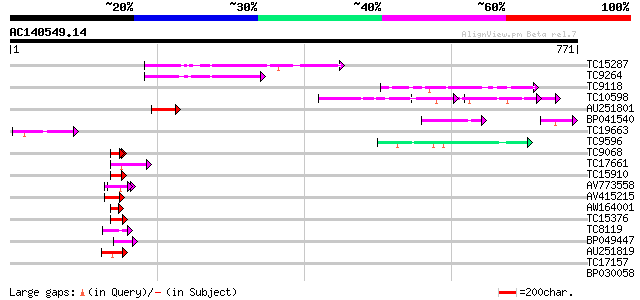
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140549.14 - phase: 0 /pseudo
(771 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC15287 similar to UP|Q9FMY5 (Q9FMY5) U2 snRNP auxiliary factor,... 108 2e-24
TC9264 similar to UP|Q9FMY5 (Q9FMY5) U2 snRNP auxiliary factor, ... 93 1e-19
TC9118 similar to UP|Q84JE4 (Q84JE4) Squamosa promoter binding l... 55 6e-08
TC10598 weakly similar to GB|AAM78036.1|21928015|AY125526 At2g01... 52 5e-07
AU251801 51 8e-07
BP041540 50 1e-06
TC19663 similar to UP|Q8H6S8 (Q8H6S8) Translation initiation fac... 50 2e-06
TC9596 similar to UP|AAR24722 (AAR24722) At5g49400, partial (66%) 48 7e-06
TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 gluc... 47 9e-06
TC17661 weakly similar to UP|Q09085 (Q09085) Hydroxyproline-rich... 47 9e-06
TC15910 similar to UP|Q9AR64 (Q9AR64) Urease accessory protein G... 47 2e-05
AV773558 45 3e-05
AV415215 44 1e-04
AW164001 43 2e-04
TC15376 weakly similar to UP|Q7SYH2 (Q7SYH2) Cystatin domain fet... 43 2e-04
TC8119 similar to UP|Q9FFF5 (Q9FFF5) Nucleic acid binding protei... 42 3e-04
BP049447 42 4e-04
AU251819 42 5e-04
TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly pr... 40 0.001
BP030058 40 0.001
>TC15287 similar to UP|Q9FMY5 (Q9FMY5) U2 snRNP auxiliary factor, small
subunit, partial (81%)
Length = 1263
Score = 108 bits (271), Expect = 2e-24
Identities = 86/289 (29%), Positives = 134/289 (45%), Gaps = 17/289 (5%)
Frame = +2
Query: 184 FGTELDKAHCPFHLKTAACRFGDRCSRVHFYPDKSCTLLIKNMYNGPGLAWEQDEGLE*L 243
FGTE D+ +CPF+ K ACR GDRCSR+H P S TLL+ NMY P + G++
Sbjct: 104 FGTEKDRVNCPFYFKIGACRHGDRCSRLHNRPSISPTLLLANMYQRPDMI---TPGVD-- 268
Query: 244 SGSSPIMA*GAMMQTNGRSLQRV*DCGSKA*KTWMHL-KFGEIVNFKVCKNGSFHLRGNV 302
PI + R +Q+ + + ++ L KFGEI VC N + H+ GNV
Sbjct: 269 PQGQPI---------DPREIQQHFEDFYE--DIFLELSKFGEIETLNVCDNLADHMIGNV 415
Query: 303 YVQYKLLDSALLAYNSVNGRYFAGKQVSCKFVNLTRWKVAICGEYMKSGYKTCSHGTACN 362
YVQ++ D A A +++GR+++G+ + +F +T ++ A C +Y ++ +C+ G CN
Sbjct: 416 YVQFREEDEAAKALAALHGRFYSGRPIIAEFSPVTDFREATCRQYEEN---SCNRGGYCN 586
Query: 363 FI----------------HCFRNPGGDYEWADSDKPPPKFWVKEMVALFGHSDNCEMSRV 406
F+ H FR + PP+ G D R
Sbjct: 587 FMHVKLIGRDLRRKLFSSHSFRIRSRSPVRRSRSRSPPR--------RRGSMDRERRHRD 742
Query: 407 HGNFSALKHSSNILETDSDREMGQLYGGRSSRRRQEDERKQRTLDEEWN 455
S + SS+ +D D + +GG +R E+ R + E+WN
Sbjct: 743 RDYDSRGRRSSDRRSSDRDGGGRRRHGGSPAREGSEERRAR---IEQWN 880
>TC9264 similar to UP|Q9FMY5 (Q9FMY5) U2 snRNP auxiliary factor, small
subunit, partial (55%)
Length = 640
Score = 93.2 bits (230), Expect = 1e-19
Identities = 59/165 (35%), Positives = 87/165 (51%), Gaps = 1/165 (0%)
Frame = +3
Query: 184 FGTELDKAHCPFHLKTAACRFGDRCSRVHFYPDKSCTLLIKNMYNGPGLAWEQDEGLE*L 243
FGTE D+ +CPF+ K ACR GDRCSR+H P S TL++ NMY P D L +
Sbjct: 153 FGTEKDRVNCPFYFKIGACRHGDRCSRLHTKPTISPTLVLSNMYQRP------DMNLNII 314
Query: 244 SGSSPIMA*GAMMQTNGRSLQRV*DCGSKA*KTWMHL-KFGEIVNFKVCKNGSFHLRGNV 302
+ P + LQ D + + L K+G+I + +C N + H+ GNV
Sbjct: 315 --TQPNQPQHQPQPLDPDKLQDHFDDFYE--DLFQELSKYGQIQSLNICDNLADHMVGNV 482
Query: 303 YVQYKLLDSALLAYNSVNGRYFAGKQVSCKFVNLTRWKVAICGEY 347
YVQYK D A A ++ GR+++G+ + F +T ++ A C +Y
Sbjct: 483 YVQYKEEDHAANALMNLTGRFYSGRPIIADFSPVTDFREATCRQY 617
>TC9118 similar to UP|Q84JE4 (Q84JE4) Squamosa promoter binding
like-protein, partial (16%)
Length = 779
Score = 54.7 bits (130), Expect = 6e-08
Identities = 56/218 (25%), Positives = 92/218 (41%), Gaps = 4/218 (1%)
Frame = +2
Query: 505 SRRHEEYSDVDRVTINRDNEKPHD-RKGRNSQKSNRDSRDWIYEAGCDEDRDGDGRKHHI 563
S RHEE + + + EK D R + S+K +R R + DRD DGRKH
Sbjct: 5 SSRHEEEEEEEE---EEEEEKREDVRDHKRSRKKHRSHRS----SHTSRDRDRDGRKH-- 157
Query: 564 SRRKGS---RHQSRDNSNVTDESESDKDRGHEEYSDVDWDTMSRENEKQHGSNGRNSQKW 620
RR S ++ R + + E H +Y D D R + ++ + + +
Sbjct: 158 KRRHSSSKDKYSYRGAKHGISDDEHQHSSHHHKY-DSSSDEKHRSSRRRQREDSMSDHEH 334
Query: 621 NRDSRYRIYEAGSDEDRHRKRHHSSQRKGSRHRSRDNSNVADESESDKDRGHEKYSDVDW 680
R++ + DE RHR RH ++H+SR + E ES+ + G S
Sbjct: 335 KHSRRHKHETSSEDERRHRSRH-------TKHKSRSHV----ERESELEEGEVMKS---- 469
Query: 681 DTMSRDNEKQHDSKGKNSQKRNGDSKYQIDEAGSDEDR 718
D + +E + S+ ++ N + +I + SDE R
Sbjct: 470 DKSQQASEVERASREPSASLSNANHGPEISDV-SDELR 580
Score = 38.1 bits (87), Expect = 0.005
Identities = 32/146 (21%), Positives = 59/146 (39%), Gaps = 1/146 (0%)
Frame = +2
Query: 622 RDSRYRIYEAGSDEDRHRKRHHSSQRKGSRHRSRDNSNVADESESDKDRGHEKYSDVDWD 681
R SR+ E +E+ KR K SR + R + + S + +DR +
Sbjct: 2 RSSRHEEEEEEEEEEEEEKREDVRDHKRSRKKHRSHRS----SHTSRDRDRDGRKHKRRH 169
Query: 682 TMSRDNEKQHDSK-GKNSQKRNGDSKYQIDEAGSDEDRHRKKHLSSQMKESRHQSSDSGY 740
+ S+D +K G + + S + ++ SDE KH SS+ ++ SD +
Sbjct: 170 SSSKDKYSYRGAKHGISDDEHQHSSHHHKYDSSSDE-----KHRSSRRRQREDSMSDHEH 334
Query: 741 GIDESDKDKGEMEAQHGYSRKSSRHK 766
K + E + + + ++HK
Sbjct: 335 KHSRRHKHETSSEDERRHRSRHTKHK 412
Score = 36.2 bits (82), Expect = 0.020
Identities = 32/152 (21%), Positives = 60/152 (39%), Gaps = 23/152 (15%)
Frame = +2
Query: 415 HSSNILETDSDREMGQLYGGRSSRRRQ-----------------EDERKQRTLDEEWNTN 457
H S+ D DR+ GR +RR +DE + + +++++
Sbjct: 107 HRSSHTSRDRDRD------GRKHKRRHSSSKDKYSYRGAKHGISDDEHQHSSHHHKYDSS 268
Query: 458 YKDNHKRKYRNKTSDSDSDKEGLEEVDRKKYHEHTRKSSFNWNKDDNSR------RHEEY 511
+ H+ R + DS SD E H+H+R+ + +D R +H+
Sbjct: 269 SDEKHRSSRRRQREDSMSDHE----------HKHSRRHKHETSSEDERRHRSRHTKHKSR 418
Query: 512 SDVDRVTINRDNEKPHDRKGRNSQKSNRDSRD 543
S V+R + + E K + + + R SR+
Sbjct: 419 SHVERESELEEGEVMKSDKSQQASEVERASRE 514
Score = 28.9 bits (63), Expect = 3.2
Identities = 26/164 (15%), Positives = 61/164 (36%), Gaps = 3/164 (1%)
Frame = +2
Query: 3 SNRKEKRKALKKLKRKERRKEIAEKERLEEEARLN---DPEEERKRILLEQQEAERIERD 59
S+R E+ + ++ + +E+R+++ + +R ++ R + +R R + + +D
Sbjct: 5 SSRHEEEEEEEEEEEEEKREDVRDHKRSRKKHRSHRSSHTSRDRDRDGRKHKRRHSSSKD 184
Query: 60 RIAFEEREKAWIIKQQQQRDLEEEQQQLSQRDLEQHESELEEEEEEDVDDDDDGRPEIIW 119
+ ++ + I + + + S D E+H S + E+ + D +
Sbjct: 185 KYSYRGAKHG--ISDDEHQHSSHHHKYDSSSD-EKHRSSRRRQREDSMSDHE-------- 331
Query: 120 LGNEIIIQKKNPNPLLLLHNHNHHHHHHHHQEQHEHHDNRPTSN 163
H H+ H H E H +R T +
Sbjct: 332 ------------------HKHSRRHKHETSSEDERRHRSRHTKH 409
Score = 28.5 bits (62), Expect = 4.2
Identities = 30/153 (19%), Positives = 51/153 (32%), Gaps = 9/153 (5%)
Frame = +2
Query: 16 KRKERRKEIAEKERLEEEARLNDPEEERKRILLEQQEAERIERDRIAFEEREKAWIIKQQ 75
+ +E E+E EEE R + + +R R + RDR + K +
Sbjct: 2 RSSRHEEEEEEEEEEEEEKREDVRDHKRSRKKHRSHRSSHTSRDRDRDGRKHKRRHSSSK 181
Query: 76 QQRDLEEEQQQLSQRDLEQHESELEEEEEEDVDDDDDGRPEIIWLGNEIIIQKKNPNPLL 135
+ + +S D QH S + D D+ R +++ +
Sbjct: 182 DKYSYRGAKHGISD-DEHQHSS---HHHKYDSSSDEKHRSS----------RRRQREDSM 319
Query: 136 LLHNHNHHHHHHH---------HQEQHEHHDNR 159
H H H H H H+ +H H +R
Sbjct: 320 SDHEHKHSRRHKHETSSEDERRHRSRHTKHKSR 418
>TC10598 weakly similar to GB|AAM78036.1|21928015|AY125526
At2g01100/F23H14.7 {Arabidopsis thaliana;}, partial
(33%)
Length = 902
Score = 51.6 bits (122), Expect = 5e-07
Identities = 48/190 (25%), Positives = 84/190 (43%), Gaps = 13/190 (6%)
Frame = +3
Query: 547 EAGCDEDRDGDGRKHHISRRKGSRHQSRDNSN---VTDESESDKDRGHEEYSDVDWDTMS 603
EA D + R+ + K + D+ + DES+ R H ++
Sbjct: 324 EAAAKAKADAEARERKLKAAKHKKRADSDSESDYDSDDESKRTSRRSHRKHKKRSHYDSD 503
Query: 604 RENEKQHGSNGRNSQKWNRDS---RYRIYEAGSDEDRHRKRHHSSQRKG--SRHRSRDNS 658
E K+ S R ++KW+ +S E+ S+E+R RKR + + SR S D+
Sbjct: 504 HEKRKEKLSK-RKTKKWSSESSDFSGDESESRSEEERRRKRKQKKKLRDQVSRSDSSDSD 680
Query: 659 NVADESESDKDRGHEKY-----SDVDWDTMSRDNEKQHDSKGKNSQKRNGDSKYQIDEAG 713
+ ADE S + R H+++ S D+ + ++ + S GK+ KR+ S+ +
Sbjct: 681 SSADE-VSKRKRQHKRHRVTKSSGSDFSSDEENSPVRKRSHGKH-HKRHRHSESSESDLS 854
Query: 714 SDEDRHRKKH 723
SD+ H+K H
Sbjct: 855 SDDIHHKKSH 884
Score = 47.4 bits (111), Expect = 9e-06
Identities = 36/135 (26%), Positives = 57/135 (41%), Gaps = 4/135 (2%)
Frame = +3
Query: 619 KWNRDSRYRIYEAGSDEDRHRKRHHSSQRKGSRHRSRDNSNVADESESDKDRGHEKYSDV 678
KW + E R RK + +K + S + + DES+ R H K+
Sbjct: 306 KWEQKLEAAAKAKADAEARERKLKAAKHKKRADSDSESDYDSDDESKRTSRRSHRKHKKR 485
Query: 679 DWDTMSRDNEKQHDSKGKNSQKRNGDSKYQID--EAGSDEDRHRKKHLSSQMKE--SRHQ 734
+ K+ SK K + + S + D E+ S+E+R RK+ ++++ SR
Sbjct: 486 SHYDSDHEKRKEKLSKRKTKKWSSESSDFSGDESESRSEEERRRKRKQKKKLRDQVSRSD 665
Query: 735 SSDSGYGIDESDKDK 749
SSDS DE K K
Sbjct: 666 SSDSDSSADEVSKRK 710
Score = 47.0 bits (110), Expect = 1e-05
Identities = 41/193 (21%), Positives = 84/193 (43%), Gaps = 3/193 (1%)
Frame = +3
Query: 421 ETDSDREMGQLYGGRSSRRRQEDERKQRTLDEEWNTNYKDNHKRKYRNKTSDSDSDKEGL 480
+ D++ +L + +R D D+E + +H++ + DSD +K
Sbjct: 342 KADAEARERKLKAAKHKKRADSDSESDYDSDDESKRTSRRSHRKHKKRSHYDSDHEKR-K 518
Query: 481 EEVDRKKYHEHTRKSSFNWNKDDNSRRHEEYSDVDRVTINRDNEKPHDRKGRNSQKSNRD 540
E++ ++K + + +SS +++ D++ R EE +R + +K D+ R+ +
Sbjct: 519 EKLSKRKTKKWSSESS-DFSGDESESRSEE----ERRRKRKQKKKLRDQVSRSDSSDSDS 683
Query: 541 SRDWIYEAGCDEDRDGDGRKHHISRRKGSRHQS-RDNSNVTDES--ESDKDRGHEEYSDV 597
S D + R ++H +++ GS S +NS V S + K H E S+
Sbjct: 684 SADEV------SKRKRQHKRHRVTKSSGSDFSSDEENSPVRKRSHGKHHKRHRHSESSES 845
Query: 598 DWDTMSRENEKQH 610
D + ++K H
Sbjct: 846 DLSSDDIHHKKSH 884
Score = 35.4 bits (80), Expect = 0.035
Identities = 30/152 (19%), Positives = 63/152 (40%)
Frame = +3
Query: 3 SNRKEKRKALKKLKRKERRKEIAEKERLEEEARLNDPEEERKRILLEQQEAERIERDRIA 62
S+ +++++ L K K K+ E ++ E E+R EEER+R +++ ++ RD+++
Sbjct: 498 SDHEKRKEKLSKRKTKKWSSESSDFSGDESESR---SEEERRR----KRKQKKKLRDQVS 656
Query: 63 FEEREKAWIIKQQQQRDLEEEQQQLSQRDLEQHESELEEEEEEDVDDDDDGRPEIIWLGN 122
+ D + ++S+R + + + D D++ P
Sbjct: 657 -----------RSDSSDSDSSADEVSKRKRQHKRHRVTKSSGSDFSSDEENSP------- 782
Query: 123 EIIIQKKNPNPLLLLHNHNHHHHHHHHQEQHE 154
++K++ H HH H H E E
Sbjct: 783 ---VRKRS---------HGKHHKRHRHSESSE 842
Score = 28.1 bits (61), Expect = 5.5
Identities = 19/93 (20%), Positives = 39/93 (41%)
Frame = +3
Query: 5 RKEKRKALKKLKRKERRKEIAEKERLEEEARLNDPEEERKRILLEQQEAERIERDRIAFE 64
R+ KRK KKL+ + R + ++ + +E + +R R+ + D
Sbjct: 609 RRRKRKQKKKLRDQVSRSDSSDSDSSADEVSKRKRQHKRHRV--TKSSGSDFSSDEENSP 782
Query: 65 EREKAWIIKQQQQRDLEEEQQQLSQRDLEQHES 97
R+++ ++ R E + LS D+ +S
Sbjct: 783 VRKRSHGKHHKRHRHSESSESDLSSDDIHHKKS 881
>AU251801
Length = 318
Score = 50.8 bits (120), Expect = 8e-07
Identities = 22/39 (56%), Positives = 26/39 (66%)
Frame = +1
Query: 194 PFHLKTAACRFGDRCSRVHFYPDKSCTLLIKNMYNGPGL 232
PF+ K ACR GDRCSR+H P S TLL+ NMY P +
Sbjct: 73 PFYFKIGACRHGDRCSRLHTKPSISPTLLLSNMYQRPDI 189
>BP041540
Length = 521
Score = 50.1 bits (118), Expect = 1e-06
Identities = 27/54 (50%), Positives = 32/54 (59%), Gaps = 4/54 (7%)
Frame = -2
Query: 722 KHLSSQMKESRHQSSDSGY----GIDESDKDKGEMEAQHGYSRKSSRHKRSGLQ 771
KH + SR+QS DS + ESD D+GEME+ H YSRKS RH R LQ
Sbjct: 520 KHHIDRRSSSRYQSRDSSHVTAESESESDNDRGEMESPHDYSRKSLRHMRYDLQ 359
Score = 49.7 bits (117), Expect = 2e-06
Identities = 34/91 (37%), Positives = 46/91 (50%), Gaps = 2/91 (2%)
Frame = -2
Query: 560 KHHISRRKGSRHQSRDNSNVT--DESESDKDRGHEEYSDVDWDTMSRENEKQHGSNGRNS 617
KHHI RR SR+QSRD+S+VT ESESD DRG E S D+ S + + N
Sbjct: 520 KHHIDRRSSSRYQSRDSSHVTAESESESDNDRGEME-SPHDYSRKSLRHMRYDLQNYCED 344
Query: 618 QKWNRDSRYRIYEAGSDEDRHRKRHHSSQRK 648
+ D +I S + R HH +++
Sbjct: 343 YENEND---QIDGGWSQRESERAAHHHQKKR 260
Score = 40.4 bits (93), Expect = 0.001
Identities = 29/88 (32%), Positives = 44/88 (49%), Gaps = 8/88 (9%)
Frame = -2
Query: 641 RHHSSQRKGSRHRSRDNSNVA--DESESDKDRGHEKYSDVDWDTMSRDNEKQ------HD 692
+HH +R SR++SRD+S+V ESESD DRG E S D+ S + + D
Sbjct: 520 KHHIDRRSSSRYQSRDSSHVTAESESESDNDRG-EMESPHDYSRKSLRHMRYDLQNYCED 344
Query: 693 SKGKNSQKRNGDSKYQIDEAGSDEDRHR 720
+ +N Q G S+ + + A + R
Sbjct: 343 YENENDQIDGGWSQRESERAAHHHQKKR 260
>TC19663 similar to UP|Q8H6S8 (Q8H6S8) Translation initiation factor,
partial (10%)
Length = 485
Score = 49.7 bits (117), Expect = 2e-06
Identities = 37/99 (37%), Positives = 54/99 (54%), Gaps = 9/99 (9%)
Frame = +3
Query: 4 NRKEKRKALKKLKRK--------ERRKEIAEKERLEEEARLNDPEEERKRILLEQQEAER 55
N + + A KKL + RRKE E+++ EEE +L EEER+R Q+E ER
Sbjct: 156 NESKTKAADKKLPKHVREMQEALARRKEAEERKKREEEEKLRKEEEERRR----QEELER 323
Query: 56 -IERDRIAFEEREKAWIIKQQQQRDLEEEQQQLSQRDLE 93
E R +EREK ++K++Q+ L +Q+ QR LE
Sbjct: 324 QAEEARRRKKEREKEKLLKKKQEGKLLTGKQKEEQRRLE 440
Score = 30.0 bits (66), Expect = 1.5
Identities = 23/99 (23%), Positives = 46/99 (46%), Gaps = 6/99 (6%)
Frame = +3
Query: 13 KKLKRKERRKEIAEKERLEEEARLNDPEEERKRILLEQQEAERIER-----DRIAFEERE 67
KK K+KE+ KE A N+ E + ++ ++ + + + E +E
Sbjct: 39 KKKKKKEKEKEKKAAAAAAGSAPANETVEVKAEVVESKKNESKTKAADKKLPKHVREMQE 218
Query: 68 KAWIIKQQQQRDLEEEQQQLSQRDLEQH-ESELEEEEEE 105
K+ ++R EE+++L + + E+ + ELE + EE
Sbjct: 219 ALARRKEAEERKKREEEEKLRKEEEERRRQEELERQAEE 335
>TC9596 similar to UP|AAR24722 (AAR24722) At5g49400, partial (66%)
Length = 1196
Score = 47.8 bits (112), Expect = 7e-06
Identities = 55/236 (23%), Positives = 91/236 (38%), Gaps = 25/236 (10%)
Frame = +2
Query: 501 KDDNSRRHEEYSDVDRVTINRDNEKP----------HDRKGRNSQKSNRDSRDWIYEAGC 550
K++ R +E + R EKP H G + W YE C
Sbjct: 302 KEEGPSRWQEKREAKRQMYLMSTEKPVKLGERIGVKHTMSGSAQCQKCYQGGHWTYE--C 475
Query: 551 DEDRDGDGRKHHISRRKGSRHQSR-------DNSNVTDESESDKD------RGHEEYSDV 597
+R R + K + + +N++V D+ E K R H SD
Sbjct: 476 KNERVYISRPSRTQQLKNPKLRVNVSVSYDLENNDVDDKEEKAKSSSKKAKRKHRSDSDS 655
Query: 598 DWDTMSRENEKQHGSNGRNSQKWNRDSRYRIYEAGSDEDRHRKRHHSSQRK--GSRHRSR 655
D+ E + GS+ ++ S + S+E+R R+R +Q+K G RH+
Sbjct: 656 GSDSEDSVFETESGSSSVTGSDYSSGSSSDSSSSDSEEERRRRRKKKTQKKKKGRRHKKY 835
Query: 656 DNSNVADESESDKDRGHEKYSDVDWDTMSRDNEKQHDSKGKNSQKRNGDSKYQIDE 711
+S+ + +SES SD D D S +++H S+ + ++ S YQ E
Sbjct: 836 SSSSESSDSES--------ASDSDSDDKSSRRKRRH-SRRH*IRLKDVRSSYQFGE 976
>TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 glucosidase
SCW10 precursor (Soluble cell wall protein 10) ,
partial (6%)
Length = 1209
Score = 47.4 bits (111), Expect = 9e-06
Identities = 14/22 (63%), Positives = 17/22 (76%)
Frame = -2
Query: 138 HNHNHHHHHHHHQEQHEHHDNR 159
H+H+HHHHHHHH H HH +R
Sbjct: 146 HHHHHHHHHHHHHHHHHHHHHR 81
Score = 45.4 bits (106), Expect = 3e-05
Identities = 13/19 (68%), Positives = 15/19 (78%)
Frame = -2
Query: 138 HNHNHHHHHHHHQEQHEHH 156
H+H+HHHHHHHH H HH
Sbjct: 149 HHHHHHHHHHHHHHHHHHH 93
Score = 38.9 bits (89), Expect = 0.003
Identities = 12/23 (52%), Positives = 14/23 (60%)
Frame = -2
Query: 134 LLLLHNHNHHHHHHHHQEQHEHH 156
L++ HHHHHHHH H HH
Sbjct: 173 LVVSCRRRHHHHHHHHHHHHHHH 105
>TC17661 weakly similar to UP|Q09085 (Q09085) Hydroxyproline-rich
glycoprotein (HRGP) (Fragment), partial (8%)
Length = 417
Score = 47.4 bits (111), Expect = 9e-06
Identities = 22/66 (33%), Positives = 29/66 (43%), Gaps = 11/66 (16%)
Frame = +2
Query: 138 HNHNHHHHHHHHQ-----------EQHEHHDNRPTSNPFPPESPSQPLHNVAQQPPNFGT 186
+NH HHHHHHHH H H D+R +++ P S + P N+ PP
Sbjct: 26 NNHRHHHHHHHHAILFTHLHPLQLLHHTHMDHRESTHSTIPHSSTHPTTNLLHLPPQTAP 205
Query: 187 ELDKAH 192
D H
Sbjct: 206 FQDPLH 223
Score = 28.9 bits (63), Expect = 3.2
Identities = 18/55 (32%), Positives = 22/55 (39%), Gaps = 4/55 (7%)
Frame = +2
Query: 132 NPLLLLHNHNHHHHHHHHQEQHEHHDNRPTSN--PFPPESP--SQPLHNVAQQPP 182
+PL LLH H H H H PT+N PP++ PLH P
Sbjct: 83 HPLQLLH-HTHMDHRESTHSTIPHSSTHPTTNLLHLPPQTAPFQDPLHQTRSTRP 244
>TC15910 similar to UP|Q9AR64 (Q9AR64) Urease accessory protein G, partial
(88%)
Length = 1061
Score = 46.6 bits (109), Expect = 2e-05
Identities = 16/23 (69%), Positives = 17/23 (73%), Gaps = 2/23 (8%)
Frame = +2
Query: 138 HNHNHHHHHHH--HQEQHEHHDN 158
HNH HHHHHHH H H+HHDN
Sbjct: 17 HNHEHHHHHHHSDHGHDHDHHDN 85
Score = 29.3 bits (64), Expect = 2.5
Identities = 10/21 (47%), Positives = 12/21 (56%), Gaps = 1/21 (4%)
Frame = +2
Query: 138 HNHNHH-HHHHHHQEQHEHHD 157
H+H+ H H H HH H H D
Sbjct: 41 HHHSDHGHDHDHHDNSHSHSD 103
>AV773558
Length = 469
Score = 45.4 bits (106), Expect = 3e-05
Identities = 21/40 (52%), Positives = 22/40 (54%), Gaps = 3/40 (7%)
Frame = +3
Query: 134 LLLLHNHN--HHHHHHHHQEQHEHHDNRPTSNPFP-PESP 170
LLLLH H+ HHHHHHH H H PT N P P P
Sbjct: 291 LLLLHPHHLCSHHHHHHHHHHHHHPHYTPTQNHHPIPHHP 410
Score = 43.1 bits (100), Expect = 2e-04
Identities = 20/46 (43%), Positives = 26/46 (56%), Gaps = 8/46 (17%)
Frame = +3
Query: 130 NPNPLLLLHNHNHHHHHHHH------QEQH--EHHDNRPTSNPFPP 167
+P+ L H+H+HHHHHHHH Q H HH + TS+P P
Sbjct: 303 HPHHLCSHHHHHHHHHHHHHPHYTPTQNHHPIPHHPH*KTSHPANP 440
Score = 27.7 bits (60), Expect = 7.2
Identities = 11/36 (30%), Positives = 16/36 (43%)
Frame = +3
Query: 146 HHHHQEQHEHHDNRPTSNPFPPESPSQPLHNVAQQP 181
H HH H HH + + P +P+Q H + P
Sbjct: 303 HPHHLCSHHHHHHHHHHHHHPHYTPTQNHHPIPHHP 410
>AV415215
Length = 430
Score = 43.9 bits (102), Expect = 1e-04
Identities = 15/28 (53%), Positives = 18/28 (63%)
Frame = +3
Query: 129 KNPNPLLLLHNHNHHHHHHHHQEQHEHH 156
+N P H+H+HHHHHHHH H HH
Sbjct: 141 QNLLPPFQTHHHHHHHHHHHHHLHHIHH 224
Score = 39.7 bits (91), Expect = 0.002
Identities = 12/20 (60%), Positives = 14/20 (70%)
Frame = +3
Query: 141 NHHHHHHHHQEQHEHHDNRP 160
+HHHHHHHH H HH + P
Sbjct: 168 HHHHHHHHHHHHHLHHIHHP 227
>AW164001
Length = 447
Score = 42.7 bits (99), Expect = 2e-04
Identities = 13/18 (72%), Positives = 15/18 (83%)
Frame = -3
Query: 138 HNHNHHHHHHHHQEQHEH 155
H+H+HHHHHHHHQ H H
Sbjct: 127 HHHHHHHHHHHHQNPHLH 74
Score = 38.5 bits (88), Expect = 0.004
Identities = 11/19 (57%), Positives = 15/19 (78%)
Frame = -3
Query: 137 LHNHNHHHHHHHHQEQHEH 155
+ +H+HHHHHHHH Q+ H
Sbjct: 136 VRHHHHHHHHHHHHHQNPH 80
Score = 36.2 bits (82), Expect = 0.020
Identities = 19/47 (40%), Positives = 23/47 (48%), Gaps = 6/47 (12%)
Frame = -3
Query: 120 LGNEIIIQKKNPNPLLLLHN------HNHHHHHHHHQEQHEHHDNRP 160
LGN I K+ +PL N H+HHHHHHHH H + P
Sbjct: 202 LGN--IGGKETIHPLACQRNQEWQVRHHHHHHHHHHHHHQNPHLHLP 68
>TC15376 weakly similar to UP|Q7SYH2 (Q7SYH2) Cystatin domain fetuin-like
protein, partial (6%)
Length = 582
Score = 42.7 bits (99), Expect = 2e-04
Identities = 12/23 (52%), Positives = 16/23 (69%)
Frame = +1
Query: 138 HNHNHHHHHHHHQEQHEHHDNRP 160
H+H H+HHHH H+ H HH + P
Sbjct: 385 HHHRHYHHHHRHRHHHRHHHHHP 453
Score = 38.9 bits (89), Expect = 0.003
Identities = 12/23 (52%), Positives = 13/23 (56%)
Frame = +1
Query: 138 HNHNHHHHHHHHQEQHEHHDNRP 160
H H HHHH H H +H HH P
Sbjct: 391 HRHYHHHHRHRHHHRHHHHHPLP 459
Score = 37.7 bits (86), Expect = 0.007
Identities = 12/31 (38%), Positives = 16/31 (50%)
Frame = +1
Query: 136 LLHNHNHHHHHHHHQEQHEHHDNRPTSNPFP 166
L +HHH H+HH +H HH +P P
Sbjct: 367 LPQRRHHHHRHYHHHHRHRHHHRHHHHHPLP 459
>TC8119 similar to UP|Q9FFF5 (Q9FFF5) Nucleic acid binding protein-like,
partial (85%)
Length = 1249
Score = 42.4 bits (98), Expect = 3e-04
Identities = 18/40 (45%), Positives = 23/40 (57%)
Frame = +1
Query: 127 QKKNPNPLLLLHNHNHHHHHHHHQEQHEHHDNRPTSNPFP 166
Q N + L+ LH H+HHHHHHHH H H + +P P
Sbjct: 91 QSPNFHSLIFLHYHHHHHHHHHH---HMHLRSLLLMHPHP 201
>BP049447
Length = 478
Score = 42.0 bits (97), Expect = 4e-04
Identities = 15/32 (46%), Positives = 19/32 (58%)
Frame = +3
Query: 142 HHHHHHHHQEQHEHHDNRPTSNPFPPESPSQP 173
HHHHHHHHQ+ + +RP P PP+ P
Sbjct: 171 HHHHHHHHQQPFSNLPSRPPPPPPPPQHHHHP 266
Score = 36.6 bits (83), Expect = 0.016
Identities = 18/58 (31%), Positives = 24/58 (41%), Gaps = 19/58 (32%)
Frame = +3
Query: 135 LLLHNHNHHHH-------------------HHHHQEQHEHHDNRPTSNPFPPESPSQP 173
LL H+H+HHHH HHHH + H +P+S P P+ P
Sbjct: 162 LLPHHHHHHHHQQPFSNLPSRPPPPPPPPQHHHHPDPKFTHHEQPSSTTQPSVRPNLP 335
Score = 35.0 bits (79), Expect = 0.045
Identities = 15/35 (42%), Positives = 17/35 (47%)
Frame = +3
Query: 141 NHHHHHHHHQEQHEHHDNRPTSNPFPPESPSQPLH 175
+HHHHHHH Q N P+ P PP P H
Sbjct: 171 HHHHHHHHQQP----FSNLPSRPPPPPPPPQHHHH 263
Score = 33.1 bits (74), Expect = 0.17
Identities = 20/63 (31%), Positives = 25/63 (38%), Gaps = 8/63 (12%)
Frame = +1
Query: 130 NPNPLLLLHNHNHHHHHHHHQEQHEHHDNRPTSN--------PFPPESPSQPLHNVAQQP 181
NP+ + L + HHHHHHH N PT N F P S L +
Sbjct: 199 NPSQISL---QDLHHHHHHHNTTTTQIQNSPTMNNHHQQLNPQFDPTSHDDFLEQMLSTL 369
Query: 182 PNF 184
P+F
Sbjct: 370 PSF 378
Score = 32.0 bits (71), Expect = 0.38
Identities = 14/33 (42%), Positives = 17/33 (51%)
Frame = +3
Query: 144 HHHHHHQEQHEHHDNRPTSNPFPPESPSQPLHN 176
HHHHHH H+ + S P PP P Q H+
Sbjct: 171 HHHHHH--HHQQPFSNLPSRPPPPPPPPQHHHH 263
>AU251819
Length = 368
Score = 41.6 bits (96), Expect = 5e-04
Identities = 17/41 (41%), Positives = 25/41 (60%), Gaps = 5/41 (12%)
Frame = +2
Query: 125 IIQKKNPNPLLLLH----NHNHH-HHHHHHQEQHEHHDNRP 160
++ +++P L+ +H NH+HH HHHHHH H HH P
Sbjct: 98 MVCRRHP*KLIRIHIPLKNHHHHRHHHHHHLYLHPHHQYHP 220
Score = 30.4 bits (67), Expect = 1.1
Identities = 9/12 (75%), Positives = 11/12 (91%)
Frame = +3
Query: 141 NHHHHHHHHQEQ 152
NHHHHH+HH+ Q
Sbjct: 54 NHHHHHYHHRLQ 89
Score = 29.3 bits (64), Expect = 2.5
Identities = 9/13 (69%), Positives = 11/13 (84%)
Frame = +3
Query: 139 NHNHHHHHHHHQE 151
NH+HHH+HH QE
Sbjct: 54 NHHHHHYHHRLQE 92
Score = 27.3 bits (59), Expect = 9.5
Identities = 9/28 (32%), Positives = 13/28 (46%)
Frame = +3
Query: 127 QKKNPNPLLLLHNHNHHHHHHHHQEQHE 154
Q +N + +HHHHH+H E
Sbjct: 9 QNQNHESFTIPGKERNHHHHHYHHRLQE 92
>TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly protein
1-like protein 3, partial (81%)
Length = 1298
Score = 40.4 bits (93), Expect = 0.001
Identities = 22/80 (27%), Positives = 40/80 (49%)
Frame = +2
Query: 37 NDPEEERKRILLEQQEAERIERDRIAFEEREKAWIIKQQQQRDLEEEQQQLSQRDLEQHE 96
+D EE + ++ + RD+I +W + +Q D E+ ++ D ++ E
Sbjct: 638 DDAVEELQNLMEHDYDIGSTIRDKII--PHAVSWFTGEAEQSDFEDIEEDDEDGDEDEDE 811
Query: 97 SELEEEEEEDVDDDDDGRPE 116
+ ++EEEED DDDD+ E
Sbjct: 812 DDDDDEEEEDDDDDDEDDEE 871
Score = 32.0 bits (71), Expect = 0.38
Identities = 21/67 (31%), Positives = 26/67 (38%), Gaps = 7/67 (10%)
Frame = -1
Query: 135 LLLHNHNHHH-------HHHHHQEQHEHHDNRPTSNPFPPESPSQPLHNVAQQPPNFGTE 187
L L H HH HHHHH H HH P ++ AQ PP T+
Sbjct: 878 LPLPRHPHHRHRHPLPLHHHHHLHLHLHHHLH--------HLLQYPQNHSAQPPP*TMTQ 723
Query: 188 LDKAHCP 194
++ CP
Sbjct: 722 HEE*FCP 702
>BP030058
Length = 455
Score = 40.4 bits (93), Expect = 0.001
Identities = 14/25 (56%), Positives = 17/25 (68%)
Frame = -2
Query: 136 LLHNHNHHHHHHHHQEQHEHHDNRP 160
LLH+ HHHHHH H +H HH + P
Sbjct: 403 LLHHPLHHHHHHCHAHRHRHHRHLP 329
Score = 37.4 bits (85), Expect = 0.009
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 5/49 (10%)
Frame = -2
Query: 137 LHNHNHH-----HHHHHHQEQHEHHDNRPTSNPFPPESPSQPLHNVAQQ 180
LH+H+HH H HH H H HH +R + P + P H+ Q+
Sbjct: 388 LHHHHHHCHAHRHRHHRHLPHHLHHLHRRLRSTPPAHRIATPCHHHHQE 242
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.131 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,226,872
Number of Sequences: 28460
Number of extensions: 288694
Number of successful extensions: 9610
Number of sequences better than 10.0: 627
Number of HSP's better than 10.0 without gapping: 3669
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6377
length of query: 771
length of database: 4,897,600
effective HSP length: 97
effective length of query: 674
effective length of database: 2,136,980
effective search space: 1440324520
effective search space used: 1440324520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC140549.14


