

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140547.5 - phase: 0
(603 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
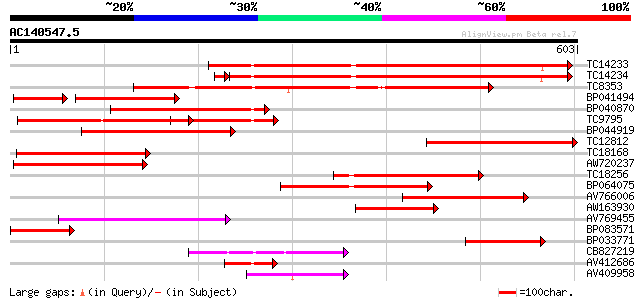
Score E
Sequences producing significant alignments: (bits) Value
TC14233 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (67%) 479 e-136
TC14234 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (66%) 446 e-128
TC8353 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa pro... 348 1e-96
BP041494 164 2e-65
BP040870 244 3e-65
TC9795 homologue to UP|Q9ATB8 (Q9ATB8) BiP-isoform D (Fragment),... 235 2e-62
BP044919 231 2e-61
TC12812 weakly similar to UP|Q8RVV9 (Q8RVV9) Heat shock protein ... 220 6e-58
TC18168 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat... 210 6e-55
AW720237 193 6e-50
TC18256 homologue to UP|Q9M4E8 (Q9M4E8) Heat shock protein 70, p... 183 6e-47
BP064075 172 2e-43
AV766006 139 1e-33
AW163930 134 4e-32
AV769455 119 1e-27
BP083571 100 1e-21
BP033771 97 6e-21
CB827219 90 9e-19
AV412686 80 9e-16
AV409958 71 4e-13
>TC14233 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (67%)
Length = 1570
Score = 479 bits (1232), Expect = e-136
Identities = 250/401 (62%), Positives = 305/401 (75%), Gaps = 14/401 (3%)
Frame = +1
Query: 212 VSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLVELFHRKYKKDLKISENFKALGRLR 271
VSL+TI+EG+F+VKAT GDTHLGG DFDN +VN V+ F RK KKD IS N +AL RLR
Sbjct: 1 VSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKD--ISGNPRALRRLR 174
Query: 272 SACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRAFFEEINKDLFKKCMETVEKCLVEV 331
+ACE+AKR LSS++QT IE+DSL GID + +TRA FEE+N DLF+KCME VEKCL +
Sbjct: 175 TACERAKRTLSSTAQTTIEIDSLYEGIDFYSPITRARFEEMNMDLFRKCMEPVEKCLRDA 354
Query: 332 KISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEVKEPCKSINPDEAVAYGAAVQAAIL 391
K+ K VH+ VLVGGST IPK+QQLL++ F KE CKSINPDEAVAYGAAVQAAIL
Sbjct: 355 KMDKRSVHDVVLVGGSTRIPKVQQLLQDFFNG----KELCKSINPDEAVAYGAAVQAAIL 522
Query: 392 NTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPKNTMIPTKKENVFSIACDNQDSLLI 451
+ EG++K++DLLLLDV P SLG+ET GGVM+VLIP+NT IPTKKE VFS DNQ +LI
Sbjct: 523 SGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLI 702
Query: 452 KVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINVCFDVDVNGILEVTAEDKTIGLKQK 511
+V+EGE +T DN LLGKFELSG PR VP I VCFD+D NGIL V+AEDKT G K K
Sbjct: 703 QVFEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNK 882
Query: 512 ITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKVKAKNLFENYVYEMRERVK------ 565
ITITN +GRLS +++ +M+++AE+YK+EDEE +KKV+AKN ENY Y MR VK
Sbjct: 883 ITITNDKGRLSKDDIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTVKDEKIAG 1062
Query: 566 --------KLEKVVEESIEWFERNQLAEIDEFEFKKQELEN 598
K+E +E++I+W E NQLAE DEFE K +ELE+
Sbjct: 1063KLDSADKTKIEDAIEQAIQWLESNQLAEADEFEDKMKELES 1185
>TC14234 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (66%)
Length = 1528
Score = 446 bits (1147), Expect(2) = e-128
Identities = 233/379 (61%), Positives = 284/379 (74%), Gaps = 14/379 (3%)
Frame = +3
Query: 234 GGIDFDNNLVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDS 293
GG DFDN +VN V+ F RK KKD IS N +AL RLR+ACE+AKR LSS++QT IE+DS
Sbjct: 48 GGEDFDNRMVNHFVQEFKRKNKKD--ISGNPRALRRLRTACERAKRTLSSTAQTTIEIDS 221
Query: 294 LCGGIDLHFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKI 353
L GID + +TRA FEE+N DLF+KCME VEKCL + K+ K VH+ VLVGGST IPK+
Sbjct: 222 LYEGIDFYSPITRARFEELNMDLFRKCMEPVEKCLRDAKMDKKSVHDVVLVGGSTRIPKV 401
Query: 354 QQLLKNMFRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLG 413
QQLL++ F KE CKSINPDEAVAYGAAVQAAIL+ EG++K++DLLLLDV P SLG
Sbjct: 402 QQLLQDFFNG----KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLG 569
Query: 414 VETKGGVMSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELS 473
+ET GGVM+VLIP+NT IPTKKE VFS DNQ +LI+VYEGE +T DN LLGKFELS
Sbjct: 570 LETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELS 749
Query: 474 GCSLVPRKVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDA 533
G PR VP I VCFD+D NGIL V+AEDKT G K KITITN +GRLS E++ +M+++A
Sbjct: 750 GIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEA 929
Query: 534 ERYKAEDEEVRKKVKAKNLFENYVYEMRERV--------------KKLEKVVEESIEWFE 579
E+YK+EDEE +KKV+AKN ENY Y MR + KK+E +E++I+W +
Sbjct: 930 EKYKSEDEEHKKKVEAKNALENYAYNMRNTIKDEKIGGKLDPADKKKIEDAIEQAIQWLD 1109
Query: 580 RNQLAEIDEFEFKKQELEN 598
NQLAE DEFE K +ELE+
Sbjct: 1110SNQLAEADEFEDKMKELES 1166
Score = 30.8 bits (68), Expect(2) = e-128
Identities = 12/16 (75%), Positives = 15/16 (93%)
Frame = +1
Query: 218 DEGMFQVKATLGDTHL 233
+EG+F+VKAT GDTHL
Sbjct: 1 EEGIFEVKATAGDTHL 48
>TC8353 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa protein,
mitochondrial precursor, partial (74%)
Length = 1752
Score = 348 bits (894), Expect = 1e-96
Identities = 201/387 (51%), Positives = 257/387 (65%), Gaps = 4/387 (1%)
Frame = +3
Query: 132 EVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYGLDKK 191
E AE YLG V+ AVITVPAYFN++ RQAT DAG+IAG +V RIINEPTAAA++YG+ K
Sbjct: 3 ETAEAYLGKSVSKAVITVPAYFNDAHRQATKDAGRIAGLDVQRIINEPTAAALSYGMTNK 182
Query: 192 MWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLVELFH 251
E + VFDLGGGTFDVS++ I G+F+VKAT GDT LGG DFDN L++ LV+ F
Sbjct: 183 -----EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVDEFK 347
Query: 252 RKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLC----GGIDLHFTVTRA 307
R DL S++ AL RLR A EKAK LSS+SQT I L + G L+ T+TR+
Sbjct: 348 RTESIDL--SKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRS 521
Query: 308 FFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEV 367
FE + L ++ + CL + IS +V E +LVGG T +PK+Q+++ +F
Sbjct: 522 KFEALVNHLIERTKAPCKNCLKDANISIKEVDEVLLVGGMTRVPKVQEVVSAIFG----- 686
Query: 368 KEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPK 427
K P K +NPDEAVA GAA+Q IL GD +++LLLLDV P SLG+ET GG+ + LI +
Sbjct: 687 KSPSKGVNPDEAVAMGAAIQGGILR--GD--VKELLLLDVTPLSLGIETLGGIFTRLINR 854
Query: 428 NTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINV 487
NT IPTKK VFS A DNQ + IKV +GE DN LG+FEL G PR +P I V
Sbjct: 855 NTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKSLGEFELVGIPPAPRGLPQIEV 1034
Query: 488 CFDVDVNGILEVTAEDKTIGLKQKITI 514
FD+D NGI+ V+A+DK+ G +Q+ITI
Sbjct: 1035TFDIDANGIVTVSAKDKSTGKEQQITI 1115
>BP041494
Length = 572
Score = 164 bits (414), Expect(2) = 2e-65
Identities = 78/110 (70%), Positives = 92/110 (82%)
Frame = +3
Query: 71 DAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVLSKL 130
DAKRLIGR+FSD +V+ D+KLWPFKV+ D+PMIVVNYK +EK F+ +EISSMVL K+
Sbjct: 243 DAKRLIGRKFSDDSVESDMKLWPFKVISGPVDRPMIVVNYKNEEKQFTAEEISSMVLIKM 422
Query: 131 KEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPT 180
+E+AE YLG V N V+TVPAYFN+SQRQAT DAG IAG NVMRIINEPT
Sbjct: 423 REIAEAYLGSTVKNVVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPT 572
Score = 102 bits (255), Expect(2) = 2e-65
Identities = 47/57 (82%), Positives = 53/57 (92%)
Frame = +1
Query: 5 KSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQL 61
+S AIGIDLGT+YSC+ VW+N+RVEIIPNDQGNR TPSYVAFTD ERLIGDAAKNQ+
Sbjct: 76 ESPAIGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDFERLIGDAAKNQM 246
>BP040870
Length = 505
Score = 244 bits (623), Expect = 3e-65
Identities = 123/169 (72%), Positives = 142/169 (83%)
Frame = +2
Query: 108 VNYKGQEKHFSPQEISSMVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKI 167
VNYKG+EK FS +EISSMVL K+KE+AE YLG + NAV+TVPAYFN+SQRQAT DAG I
Sbjct: 5 VNYKGEEKQFSAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVI 184
Query: 168 AGFNVMRIINEPTAAAIAYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKAT 227
+G NVMRIINEPTAAAIAYGLDKK GEKNVL+FDLGGGTFDVSL+TI+EG+F+VK+T
Sbjct: 185 SGLNVMRIINEPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKST 364
Query: 228 LGDTHLGGIDFDNNLVNRLVELFHRKYKKDLKISENFKALGRLRSACEK 276
GDTHLGG DFDN +VN V+ F RK KKD IS N +AL RLR+ACE+
Sbjct: 365 AGDTHLGGEDFDNRMVNHFVQEFKRKNKKD--ISGNARALRRLRTACER 505
>TC9795 homologue to UP|Q9ATB8 (Q9ATB8) BiP-isoform D (Fragment), partial
(63%)
Length = 981
Score = 235 bits (599), Expect = 2e-62
Identities = 116/188 (61%), Positives = 146/188 (76%), Gaps = 1/188 (0%)
Frame = +1
Query: 9 IGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHNT 68
IGIDLGT+YSC+ V+RN VEII NDQGNR+TPS+VAF D ERLIG+AAKNQ A P T
Sbjct: 154 IGIDLGTTYSCVGVYRNGHVEIIANDQGNRITPSWVAFNDGERLIGEAAKNQFAVNPERT 333
Query: 69 VFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYK-GQEKHFSPQEISSMVL 127
+FD KRLIGR+F D+ VQ+D+KL P+K+V N KP I V K G+ K FSP+E+S+M+L
Sbjct: 334 IFDVKRLIGRKFEDKEVQRDMKLVPYKIV-NKDGKPYIQVKIKDGETKVFSPEEVSAMIL 510
Query: 128 SKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYG 187
+K+KE AE +LG ++N+AV+TVPAYFN++QRQAT DAG IAG NV RIINEPT
Sbjct: 511 TKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTCCCHCLW 690
Query: 188 LDKKMWRE 195
+ ++ WRE
Sbjct: 691 IGQERWRE 714
Score = 125 bits (314), Expect = 2e-29
Identities = 66/115 (57%), Positives = 85/115 (73%)
Frame = +2
Query: 172 VMRIINEPTAAAIAYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDT 231
++ + P AAAIAYGLDKK GEKN+LVFDLGGGTFDVS++TID G+F+V AT GDT
Sbjct: 644 LLELSMNPPAAAIAYGLDKK---GGEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDT 814
Query: 232 HLGGIDFDNNLVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQ 286
HLGG DFD ++ ++L +K+ KD IS++ +ALG+LR E+AKR LSS Q
Sbjct: 815 HLGGEDFDQRIMEYFIKLIKKKHGKD--ISKDNRALGKLRREAERAKRALSSQHQ 973
>BP044919
Length = 519
Score = 231 bits (590), Expect = 2e-61
Identities = 111/164 (67%), Positives = 134/164 (81%)
Frame = +3
Query: 77 GRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVLSKLKEVAET 136
GR++SD +Q+D+KLWPFKV+ DKPMIVV +KG+EK P+EISSMVLSK+KE+AE
Sbjct: 27 GRKYSDSVIQEDVKLWPFKVIAGTDDKPMIVVKHKGEEKKLVPEEISSMVLSKMKEIAEA 206
Query: 137 YLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYGLDKKMWREG 196
+L V NAV+TVPAYFN++QR+AT DAG IAG NVMRIINEPTAAA+AYGL K+ G
Sbjct: 207 FLESSVKNAVVTVPAYFNDAQRKATKDAGTIAGLNVMRIINEPTAAALAYGLQKRANLVG 386
Query: 197 EKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDN 240
E+NV +FDLGGGTFDVSL+TI + F+VKAT GDTHLG DFDN
Sbjct: 387 ERNVFIFDLGGGTFDVSLLTIKDDNFEVKATSGDTHLGXKDFDN 518
>TC12812 weakly similar to UP|Q8RVV9 (Q8RVV9) Heat shock protein 70
(Fragment), partial (48%)
Length = 746
Score = 220 bits (560), Expect = 6e-58
Identities = 111/160 (69%), Positives = 129/160 (80%)
Frame = +1
Query: 444 DNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINVCFDVDVNGILEVTAED 503
DNQ S+ IKVYEGE AK + NF LGKFELSG S P+ INVCFDVD +GI+EV+AED
Sbjct: 1 DNQTSVWIKVYEGEGAKADKNFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSAED 180
Query: 504 KTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKVKAKNLFENYVYEMRER 563
++ LK+ ITITN+ GRLS EEM RM+RDA +YKAEDEEVRKKVKAKN ENY YE R+R
Sbjct: 181 MSLRLKKTITITNRLGRLSQEEMSRMVRDAAQYKAEDEEVRKKVKAKNSLENYAYEARDR 360
Query: 564 VKKLEKVVEESIEWFERNQLAEIDEFEFKKQELENSMKFL 603
VKKLEK+VEE IEW +RNQLAE +EFE+KKQELE M F+
Sbjct: 361 VKKLEKMVEEVIEWLDRNQLAEAEEFEYKKQELEKDMPFV 480
>TC18168 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat shock
cognate protein 70, partial (23%)
Length = 508
Score = 210 bits (534), Expect = 6e-55
Identities = 101/142 (71%), Positives = 120/142 (84%)
Frame = +2
Query: 8 AIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHN 67
AIGIDLGT+YSC+ VW+++RVEII NDQGNR TPSYVAFTDTERLIGDAAKNQ+A P N
Sbjct: 83 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAMNPIN 262
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
TVFDAKRLIGRR SD +VQ D+KLWPFKV +KPMI V+YKG++K F+ +EISSMVL
Sbjct: 263 TVFDAKRLIGRRVSDASVQSDMKLWPFKVNSGPAEKPMIQVSYKGEDKQFAAEEISSMVL 442
Query: 128 SKLKEVAETYLGHEVNNAVITV 149
K++E+AE YLG + NAV+TV
Sbjct: 443 MKMREIAEAYLGSTIKNAVVTV 508
>AW720237
Length = 459
Score = 193 bits (491), Expect = 6e-50
Identities = 92/142 (64%), Positives = 112/142 (78%)
Frame = +3
Query: 5 KSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKY 64
++ AIGIDLGT+YSC+ VW N+RVEII NDQGNR TPSYV FT++ERLIGDAAKNQ+A+
Sbjct: 33 QATAIGIDLGTTYSCVGVWMNDRVEIIANDQGNRTTPSYVGFTESERLIGDAAKNQVARN 212
Query: 65 PHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISS 124
P NTVFDAKRLIGR+F D VQ+DIKLWPFKV KP+I V YKG+ K F +++SS
Sbjct: 213 PINTVFDAKRLIGRKFDDPIVQKDIKLWPFKVEAGPDGKPLIAVKYKGETKKFHAEQVSS 392
Query: 125 MVLSKLKEVAETYLGHEVNNAV 146
MVL+K+KE AE YL + + V
Sbjct: 393 MVLTKMKETAEAYLNKTIKDIV 458
>TC18256 homologue to UP|Q9M4E8 (Q9M4E8) Heat shock protein 70, partial
(24%)
Length = 476
Score = 183 bits (465), Expect = 6e-47
Identities = 96/160 (60%), Positives = 119/160 (74%)
Frame = +3
Query: 345 GGSTTIPKIQQLLKNMFRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLL 404
GGST IPK+QQLLK+ F E KEP K +NPDEAVAYGAAVQ +IL+ EG ++ +D+LL
Sbjct: 3 GGSTRIPKVQQLLKDYF----EGKEPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILL 170
Query: 405 LDVMPFSLGVETKGGVMSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDN 464
LDV P +LG+ET GGVM+ LIP+NT+IPTKK VF+ D Q ++ I+V+EGE + T+D
Sbjct: 171 LDVAPLTLGIETVGGVMTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDC 350
Query: 465 FLLGKFELSGCSLVPRKVPNINVCFDVDVNGILEVTAEDK 504
LLGKF+LSG PR P I V F+VD NGIL V AEDK
Sbjct: 351 RLLGKFDLSGIPPAPRGTPQIEVTFEVDANGILNVKAEDK 470
>BP064075
Length = 554
Score = 172 bits (435), Expect = 2e-43
Identities = 87/161 (54%), Positives = 117/161 (72%)
Frame = -2
Query: 289 IELDSLCGGIDLHFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGST 348
IELD L GID + ++TRA FEE+NKD F+KCME VEKC+++ K+ KS +H+ VLVGGS+
Sbjct: 550 IELDFLYQGIDFYSSITRAKFEELNKDYFQKCMELVEKCVIDSKMDKSNIHDVVLVGGSS 371
Query: 349 TIPKIQQLLKNMFRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVM 408
IPK++QLL + F K+ C INPDEAVA+GAAV A+IL+ E +K++ LLL +V
Sbjct: 370 RIPKVRQLLMDFF----GRKDLCNRINPDEAVAHGAAVHASILSGEFSEKVQSLLLREVT 203
Query: 409 PFSLGVETKGGVMSVLIPKNTMIPTKKENVFSIACDNQDSL 449
P SLG+E GG+M +IP+NTMIPT ++VF+ NQ L
Sbjct: 202 PLSLGLEKHGGIMETIIPRNTMIPTTMDHVFTTHFHNQPYL 80
>AV766006
Length = 408
Score = 139 bits (350), Expect = 1e-33
Identities = 70/134 (52%), Positives = 93/134 (69%)
Frame = -1
Query: 418 GGVMSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSL 477
GGVMS +I +NT+IPT+K F+ D Q ++ I+VYEGE A T+DN LGKF+L+G
Sbjct: 405 GGVMSKIIERNTVIPTRKTQTFTTYQDKQTTVSIQVYEGERAMTKDNRQLGKFDLNGIPA 226
Query: 478 VPRKVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYK 537
R P I V F+VD NGIL V+AEDK K+ ITITN +GRLS EE+ RM+++AE +
Sbjct: 225 AARGTPQIEVTFEVDANGILAVSAEDKASSAKEAITITNDKGRLSKEEIERMVKEAEEFA 46
Query: 538 AEDEEVRKKVKAKN 551
D EV++KV+ KN
Sbjct: 45 DADAEVKRKVEGKN 4
>AW163930
Length = 359
Score = 134 bits (337), Expect = 4e-32
Identities = 67/89 (75%), Positives = 77/89 (86%)
Frame = +1
Query: 368 KEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPK 427
KE CKSINPDEAVAYGAAVQAAIL+ EG++K++DLLLLDV P SLG+ET GGVM+VLIP+
Sbjct: 52 KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPR 231
Query: 428 NTMIPTKKENVFSIACDNQDSLLIKVYEG 456
NT IPTKKE VFS DNQ +LI+VYEG
Sbjct: 232 NTTIPTKKEQVFSTYSDNQPGVLIQVYEG 318
>AV769455
Length = 556
Score = 119 bits (298), Expect = 1e-27
Identities = 67/184 (36%), Positives = 101/184 (54%), Gaps = 1/184 (0%)
Frame = +1
Query: 53 IGDAAKNQLAKYPHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKG 112
+G A + P N++ KRLIGR+F+D +Q+D+K PF V P+I Y G
Sbjct: 4 LGTAGASSTMMNPKNSISQLKRLIGRQFADPELQRDLKSLPFAVTEGPDGFPLIHARYLG 183
Query: 113 QEKHFSPQEISSMVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNV 172
+ + F+P ++ M+LS LKE+A+ L V + I +P YF + QR+A DA IAG +
Sbjct: 184 ETRTFTPTQVFGMMLSNLKEIAQKNLNAAVVDCCIGIPIYFTDLQRRAVLDAATIAGLHP 363
Query: 173 MRIINEPTAAAIAYGLDKKMWREGEK-NVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDT 231
+R+ +E TA A+AYG+ K E + NV D+G + V + +G +V A D
Sbjct: 364 LRLFHETTATALAYGIYKTDLPENDPLNVAFVDVGHASMQVCIAGYKKGQLKVLAHSYDR 543
Query: 232 HLGG 235
LGG
Sbjct: 544 SLGG 555
>BP083571
Length = 423
Score = 99.8 bits (247), Expect = 1e-21
Identities = 51/72 (70%), Positives = 60/72 (82%), Gaps = 3/72 (4%)
Frame = +1
Query: 1 MATR-KSKAIGIDLGTSYSCIAVWR--NNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAA 57
MAT+ + AIGIDLGT+YSC+AVW+ N+R EII N+QGNR TPS+VAFTDT+RLIGDAA
Sbjct: 145 MATKYEGPAIGIDLGTTYSCVAVWQEQNDRAEIIHNEQGNRTTPSFVAFTDTQRLIGDAA 324
Query: 58 KNQLAKYPHNTV 69
KNQ A P NTV
Sbjct: 325 KNQAATNPTNTV 360
>BP033771
Length = 513
Score = 97.4 bits (241), Expect = 6e-21
Identities = 41/85 (48%), Positives = 66/85 (77%)
Frame = -2
Query: 485 INVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVR 544
+NVCFD+D NGIL V+AE++T G+K IT+TN GRLS E++ R++++AE+++AED
Sbjct: 509 LNVCFDLDANGILNVSAEEETTGIKNGITVTNDNGRLSSEQIMRLIQEAEKHRAEDRMYE 330
Query: 545 KKVKAKNLFENYVYEMRERVKKLEK 569
KKV+A N ++YVY++R +K +++
Sbjct: 329 KKVEAMNALDDYVYKLRNAIKDIDR 255
>CB827219
Length = 544
Score = 90.1 bits (222), Expect = 9e-19
Identities = 52/170 (30%), Positives = 95/170 (55%)
Frame = +3
Query: 191 KMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLVELF 250
++W G + +L F++G G DV++ G+ Q+KA G T +GG D N+++ L+
Sbjct: 15 RIWAVGVRKLLYFNMGAGYCDVAVTVTAGGVSQIKALAGST-IGGEDLLQNMMHHLLPDC 191
Query: 251 HRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRAFFE 310
+K + K++G LR A A LSS +++D L G+ ++ + FE
Sbjct: 192 ENIFKNHGV--KEIKSMGLLRVATLDAIHRLSSQKSVQVDVD-LGDGLKINKVIDCEEFE 362
Query: 311 EINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNM 360
E+NK++F+KC + +CL + KI V++ ++VGG + IP+++ L+ N+
Sbjct: 363 EVNKNVFEKCESLIIQCLQDSKIEVEDVNDVIIVGGCSYIPRVKNLVTNI 512
>AV412686
Length = 166
Score = 80.1 bits (196), Expect = 9e-16
Identities = 40/57 (70%), Positives = 48/57 (84%)
Frame = +2
Query: 229 GDTHLGGIDFDNNLVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSS 285
GDT+LGG+DFDNNLVN LV+ F RK+ KD I+EN K+L RLR ACEKAKR+LSS+S
Sbjct: 2 GDTYLGGVDFDNNLVNFLVDFFQRKHNKD--ITENVKSLRRLRLACEKAKRILSSTS 166
>AV409958
Length = 389
Score = 71.2 bits (173), Expect = 4e-13
Identities = 44/112 (39%), Positives = 63/112 (55%), Gaps = 4/112 (3%)
Frame = +2
Query: 253 KYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGID----LHFTVTRAF 308
K + + + ++ +AL RL EKAK LSS +QT I L + D + T+TRA
Sbjct: 32 KRNEGIDLLKDKQALQRLTETAEKAKMELSSLTQTNISLPFITATADGPKHIETTLTRAK 211
Query: 309 FEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNM 360
FEE+ DL + VE L + K+S + E +LVGGST IP +Q+L+K M
Sbjct: 212 FEELCSDLLDRLRTPVENSLRDAKLSFKDLDEVILVGGSTRIPAVQELVKKM 367
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.135 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,592,806
Number of Sequences: 28460
Number of extensions: 101702
Number of successful extensions: 511
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 480
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 487
length of query: 603
length of database: 4,897,600
effective HSP length: 96
effective length of query: 507
effective length of database: 2,165,440
effective search space: 1097878080
effective search space used: 1097878080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC140547.5


