

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140030.4 - phase: 0 /pseudo
(148 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
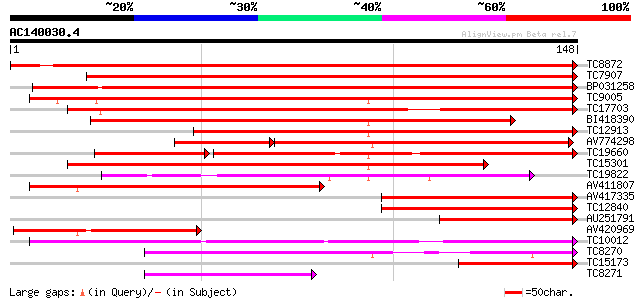
Score E
Sequences producing significant alignments: (bits) Value
TC8872 homologue to UP|Q8L5G5 (Q8L5G5) Alpha-expansin 3, partial... 287 5e-79
TC7907 homologue to UP|O81133 (O81133) Expansin, partial (97%) 249 1e-67
BP031258 242 2e-65
TC9005 similar to UP|Q84L76 (Q84L76) Expansin precursor, partial... 230 7e-62
TC17703 homologue to UP|CAC19183 (CAC19183) Expansin, partial (82%) 216 2e-57
BI418390 195 3e-51
TC12913 homologue to UP|Q84L76 (Q84L76) Expansin precursor, part... 169 1e-43
AV774298 128 1e-41
TC19660 similar to GB|BAB09383.1|9758857|AB010694 expansin-like ... 112 2e-32
TC15301 similar to UP|EX13_ARATH (Q9M9P0) Alpha-expansin 13 prec... 128 4e-31
TC19822 similar to UP|EX13_ARATH (Q9M9P0) Alpha-expansin 13 prec... 105 2e-24
AV411807 97 1e-21
AV417335 88 5e-19
TC12840 similar to PIR|T02010|T02010 expansin homolog T15B16.16 ... 86 2e-18
AU251791 71 9e-14
AV420969 67 2e-12
TC10012 weakly similar to UP|EXR1_ARATH (O23547) Expansin-relate... 56 3e-09
TC8270 similar to UP|Q7XHJ2 (Q7XHJ2) Expansin-like protein (Frag... 54 1e-08
TC15173 similar to UP|AAR09168 (AAR09168) Alpha-expansin 1, part... 47 1e-06
TC8271 weakly similar to UP|EXL2_ARATH (Q9SVE5) Expansin-like 2 ... 40 2e-04
>TC8872 homologue to UP|Q8L5G5 (Q8L5G5) Alpha-expansin 3, partial (82%)
Length = 729
Score = 287 bits (735), Expect = 5e-79
Identities = 131/148 (88%), Positives = 137/148 (92%)
Frame = +2
Query: 1 MAFIGLVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNT 60
MA IG +L LTMF+S YAYGGGWT+AHATFYGG DASGTMGGACGYGNLYSQGYGTNT
Sbjct: 89 MALIGFLL---LTMFTSAYAYGGGWTDAHATFYGGGDASGTMGGACGYGNLYSQGYGTNT 259
Query: 61 AALSTALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPL 120
AALSTA+FNNGLSCGSCYE+RC NDHRWCLPGSIVVTATNFCPPNNALPNN GGWCNPPL
Sbjct: 260 AALSTAMFNNGLSCGSCYEVRCVNDHRWCLPGSIVVTATNFCPPNNALPNNAGGWCNPPL 439
Query: 121 QHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
HFDLAQPVFLRIAQYKAGIVPV +RRV
Sbjct: 440 HHFDLAQPVFLRIAQYKAGIVPVSYRRV 523
>TC7907 homologue to UP|O81133 (O81133) Expansin, partial (97%)
Length = 1313
Score = 249 bits (637), Expect = 1e-67
Identities = 110/128 (85%), Positives = 118/128 (91%)
Frame = +2
Query: 21 YGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEI 80
YGGGW HATFYGG DASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYE+
Sbjct: 158 YGGGWQGGHATFYGGGDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEM 337
Query: 81 RCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAGI 140
RC +D RWCLPGSI+VTATNFCPPN AL N++GGWCNPPLQHFDLA+P FL+IAQYKAGI
Sbjct: 338 RCDSDPRWCLPGSILVTATNFCPPNYALSNSNGGWCNPPLQHFDLAEPAFLKIAQYKAGI 517
Query: 141 VPVDFRRV 148
VP+ FRRV
Sbjct: 518 VPISFRRV 541
>BP031258
Length = 526
Score = 242 bits (617), Expect = 2e-65
Identities = 110/142 (77%), Positives = 123/142 (86%)
Frame = +1
Query: 7 VLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTA 66
VL + M + YGG W +AHATFYGG DASGTMGGACGYGNLYSQGYGT+TAALSTA
Sbjct: 76 VLFIGMNMQGAAADYGG-WESAHATFYGGGDASGTMGGACGYGNLYSQGYGTDTAALSTA 252
Query: 67 LFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLA 126
LFNNGLSCGSCYE+RC +D RWC PGSI+VTATNFCPPN +L NN+GGWCNPPLQHFD+A
Sbjct: 253 LFNNGLSCGSCYEMRCDDDPRWCKPGSIIVTATNFCPPNPSLANNNGGWCNPPLQHFDMA 432
Query: 127 QPVFLRIAQYKAGIVPVDFRRV 148
+P FL+IAQY+AGIVPV FRRV
Sbjct: 433 EPAFLQIAQYRAGIVPVAFRRV 498
>TC9005 similar to UP|Q84L76 (Q84L76) Expansin precursor, partial (92%)
Length = 1277
Score = 230 bits (587), Expect = 7e-62
Identities = 109/154 (70%), Positives = 126/154 (81%), Gaps = 11/154 (7%)
Frame = +1
Query: 6 LVLVCS---LTMFSSVYAY------GGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGY 56
+VL+C+ +T+F + A GG W +AHATFYGGSDASGTMGGACGYGNLYSQGY
Sbjct: 55 MVLLCAASLVTLFVTTSARIPGVYTGGAWQSAHATFYGGSDASGTMGGACGYGNLYSQGY 234
Query: 57 GTNTAALSTALFNNGLSCGSCYEIRCANDHRWCLPG--SIVVTATNFCPPNNALPNNDGG 114
G NTAALSTALFNNGLSCG+C+EI+C D RWC PG SI+VTATNFCPPN A P++DGG
Sbjct: 235 GVNTAALSTALFNNGLSCGACFEIKCDQDPRWCNPGSPSIIVTATNFCPPNLAQPSDDGG 414
Query: 115 WCNPPLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
WCNPP HFDLA P+FL+IAQY+AGIVPV +RRV
Sbjct: 415 WCNPPRTHFDLAMPMFLKIAQYRAGIVPVTYRRV 516
>TC17703 homologue to UP|CAC19183 (CAC19183) Expansin, partial (82%)
Length = 687
Score = 216 bits (549), Expect = 2e-57
Identities = 102/134 (76%), Positives = 112/134 (83%), Gaps = 1/134 (0%)
Frame = +1
Query: 16 SSVYAYG-GGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSC 74
S V YG GGWTNAHATFYGGSDASGTMGGACGYGNL SQGYGTNTAALSTALFN+GLSC
Sbjct: 52 SHVNGYGDGGWTNAHATFYGGSDASGTMGGACGYGNLISQGYGTNTAALSTALFNSGLSC 231
Query: 75 GSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIA 134
G+C++I+C ND +WCLPGSIVVTATN CPP GG C+PP HFDL+QP+F IA
Sbjct: 232 GACFQIKCVNDPQWCLPGSIVVTATNVCPP--------GGGCDPPNHHFDLSQPIFQHIA 387
Query: 135 QYKAGIVPVDFRRV 148
QYKAGIVPV +RRV
Sbjct: 388 QYKAGIVPVAYRRV 429
>BI418390
Length = 471
Score = 195 bits (495), Expect = 3e-51
Identities = 87/113 (76%), Positives = 96/113 (83%), Gaps = 2/113 (1%)
Frame = +1
Query: 22 GGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIR 81
GG W AHATFYGGSDASGTMGGACGYGNLYSQGYG NTAALSTALFNNGLSCG+C+E++
Sbjct: 133 GGAWQTAHATFYGGSDASGTMGGACGYGNLYSQGYGVNTAALSTALFNNGLSCGACFELK 312
Query: 82 CANDHRWCLPG--SIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLR 132
CAND WC G SI VTATNFCPPN A +++GGWCNPP HFDLA P+FL+
Sbjct: 313 CANDPSWCHAGSPSIFVTATNFCPPNFAQASDNGGWCNPPRPHFDLAMPMFLK 471
>TC12913 homologue to UP|Q84L76 (Q84L76) Expansin precursor, partial (78%)
Length = 606
Score = 169 bits (429), Expect = 1e-43
Identities = 76/102 (74%), Positives = 89/102 (86%), Gaps = 2/102 (1%)
Frame = +2
Query: 49 GNLYSQGYGTNTAALSTALFNNGLSCGSCYEIRCANDHRWCLPG--SIVVTATNFCPPNN 106
GNLY+QGYG NTAALSTALFNNGLSCG+C+E++C D RWC PG SI+VTATNFCPPN
Sbjct: 2 GNLYTQGYGVNTAALSTALFNNGLSCGACFEMKCDQDPRWCNPGNPSILVTATNFCPPNF 181
Query: 107 ALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
A P+++GGWCNPP HFDLA P+FL+IAQY+AGIVPV +RRV
Sbjct: 182 AQPSDNGGWCNPPRPHFDLAMPMFLKIAQYRAGIVPVAYRRV 307
>AV774298
Length = 482
Score = 128 bits (321), Expect(2) = 1e-41
Identities = 55/80 (68%), Positives = 68/80 (84%), Gaps = 2/80 (2%)
Frame = +2
Query: 70 NGLSCGSCYEIRCANDHRWCLPGS--IVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQ 127
+GLSCG+C+EI+CAND +WC GS I +TATNFCPPN ALPN++GGWCNPP FDLA
Sbjct: 80 SGLSCGACFEIKCANDKQWCHSGSPSIFITATNFCPPNYALPNDNGGWCNPPRPDFDLAM 259
Query: 128 PVFLRIAQYKAGIVPVDFRR 147
P+FL+IA+Y+AGIVPV +RR
Sbjct: 260 PMFLKIAEYRAGIVPVAYRR 319
Score = 56.6 bits (135), Expect(2) = 1e-41
Identities = 25/26 (96%), Positives = 25/26 (96%)
Frame = +1
Query: 44 GACGYGNLYSQGYGTNTAALSTALFN 69
GACGYGNLYSQGYG NTAALSTALFN
Sbjct: 1 GACGYGNLYSQGYGVNTAALSTALFN 78
>TC19660 similar to GB|BAB09383.1|9758857|AB010694 expansin-like protein
{Arabidopsis thaliana;} , partial (66%)
Length = 652
Score = 112 bits (281), Expect(2) = 2e-32
Identities = 57/97 (58%), Positives = 68/97 (69%), Gaps = 2/97 (2%)
Frame = +2
Query: 54 QGYGTNTAALSTALFNNGLSCGSCYEIRCANDHRWCLPG--SIVVTATNFCPPNNALPNN 111
QGYG +T ALSTALFNNGL+CG+C EI+C N WC S++VTATNFCPPN
Sbjct: 200 QGYGLHTTALSTALFNNGLACGACIEIKCVNS-AWCRKDAPSVLVTATNFCPPN--YTKT 370
Query: 112 DGGWCNPPLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
G CNPP +HFDL +F +IA Y+AG+VPV RRV
Sbjct: 371 VGVRCNPPQKHFDLTVFMFTKIAWYRAGVVPVQARRV 481
Score = 40.8 bits (94), Expect(2) = 2e-32
Identities = 18/30 (60%), Positives = 21/30 (70%)
Frame = +1
Query: 23 GGWTNAHATFYGGSDASGTMGGACGYGNLY 52
GG +A ATFYG GTM GACGYG+L+
Sbjct: 106 GGGFDARATFYGDFKGIGTMQGACGYGDLW 195
>TC15301 similar to UP|EX13_ARATH (Q9M9P0) Alpha-expansin 13 precursor
(At-EXP13) (AtEx13) (Ath-ExpAlpha-1.22), partial (45%)
Length = 507
Score = 128 bits (322), Expect = 4e-31
Identities = 59/111 (53%), Positives = 70/111 (62%), Gaps = 1/111 (0%)
Frame = +1
Query: 16 SSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCG 75
SS ++ W +A AT+Y +D +GGACGYG+L GYG TA LS LF G CG
Sbjct: 166 SSSWSSLSEWRSARATYYAAADPRDAVGGACGYGDLLKTGYGMATAGLSETLFERGQICG 345
Query: 76 SCYEIRCANDHRWCLPG-SIVVTATNFCPPNNALPNNDGGWCNPPLQHFDL 125
+C+E+RC D RWCLPG SI+VTATNFC PN GG CNPP HF L
Sbjct: 346 ACFELRCVEDSRWCLPGTSIIVTATNFCAPNYGFTAEGGGHCNPPNNHFVL 498
>TC19822 similar to UP|EX13_ARATH (Q9M9P0) Alpha-expansin 13 precursor
(At-EXP13) (AtEx13) (Ath-ExpAlpha-1.22), partial (27%)
Length = 532
Score = 105 bits (263), Expect = 2e-24
Identities = 61/123 (49%), Positives = 73/123 (58%), Gaps = 10/123 (8%)
Frame = -1
Query: 25 WTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIRC-- 82
W A AT Y + S ++ GACGYGN GYGT TAALS ALF G CG+C+E+RC
Sbjct: 355 WLPATATHYTAT-TSDSVDGACGYGN----GYGTATAALSQALFGRGQICGACFELRCVE 191
Query: 83 -----ANDHRWCLPG--SIVVTATNFCPPNNAL-PNNDGGWCNPPLQHFDLAQPVFLRIA 134
A D RWC+ S+VVTAT+FC PN + GG CNPP QHF L F +IA
Sbjct: 190 EESATAFDRRWCVSSSTSVVVTATDFCAPNYGFDAESRGGRCNPPKQHFVLPVEAFEKIA 11
Query: 135 QYK 137
+K
Sbjct: 10 IWK 2
>AV411807
Length = 401
Score = 97.1 bits (240), Expect = 1e-21
Identities = 45/84 (53%), Positives = 57/84 (67%), Gaps = 7/84 (8%)
Frame = +2
Query: 6 LVLVCSLTMFS-------SVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGT 58
+ L ++T+FS +V W AHATFYG AS TMGGACGYGNL+ GYGT
Sbjct: 89 MFLTFTMTLFSFMGQPALAVVFRPSKWALAHATFYGDETASETMGGACGYGNLFQSGYGT 268
Query: 59 NTAALSTALFNNGLSCGSCYEIRC 82
+T ALS+ LFNNG +CG+CY+I+C
Sbjct: 269 DTVALSSTLFNNGYACGTCYQIKC 340
>AV417335
Length = 428
Score = 88.2 bits (217), Expect = 5e-19
Identities = 37/51 (72%), Positives = 44/51 (85%)
Frame = +3
Query: 98 ATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
ATNFCPPN ALPNN+GGWCNPPL+HFD+AQP + +I Y+ GIVPV F+RV
Sbjct: 3 ATNFCPPNFALPNNNGGWCNPPLKHFDMAQPAWEKIGIYRGGIVPVLFQRV 155
>TC12840 similar to PIR|T02010|T02010 expansin homolog T15B16.16 -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(60%)
Length = 648
Score = 86.3 bits (212), Expect = 2e-18
Identities = 36/51 (70%), Positives = 43/51 (83%)
Frame = +3
Query: 98 ATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
ATNFCPPN ALPN++GGWCNPP HFD++QP F IA+Y+AGIVP+ RRV
Sbjct: 15 ATNFCPPNLALPNDNGGWCNPPRPHFDMSQPAFETIAKYRAGIVPIFHRRV 167
>AU251791
Length = 343
Score = 70.9 bits (172), Expect = 9e-14
Identities = 30/36 (83%), Positives = 33/36 (91%)
Frame = +1
Query: 113 GGWCNPPLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
GGWCNPPLQHFDL+QP F +IAQYKAGIVPV +RRV
Sbjct: 4 GGWCNPPLQHFDLSQPAFQQIAQYKAGIVPVAYRRV 111
>AV420969
Length = 219
Score = 66.6 bits (161), Expect = 2e-12
Identities = 34/59 (57%), Positives = 40/59 (67%), Gaps = 10/59 (16%)
Frame = +1
Query: 2 AFIGLVLVCSLTMFS----------SVYAYGGGWTNAHATFYGGSDASGTMGGACGYGN 50
A G+VL+C ++ S VY+ GG W +AHATFYGGSDASGTMGGACGYGN
Sbjct: 46 AMAGIVLLCISSLVSLFAATTARIPGVYS-GGPWQSAHATFYGGSDASGTMGGACGYGN 219
>TC10012 weakly similar to UP|EXR1_ARATH (O23547) Expansin-related protein 1
precursor (At-EXPR1) (Ath-ExpBeta-3.1), partial (66%)
Length = 687
Score = 55.8 bits (133), Expect = 3e-09
Identities = 38/143 (26%), Positives = 64/143 (44%)
Frame = +2
Query: 6 LVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALST 65
L L+C + + + +T + AT+YG D G GACG+G Y + A +
Sbjct: 92 LGLICVMLLLPVLCTSQDTFTCSRATYYGSPDCYGNPRGACGFGE-YGKIVNDGNVAGVS 268
Query: 66 ALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDL 125
L+ NG CG+CY++RC ++C V T+F + + P + L
Sbjct: 269 WLWKNGSGCGACYQVRC-KIPQFCDDYGAYVVVTDFGVGDRT------DFVMSPRGYSRL 427
Query: 126 AQPVFLRIAQYKAGIVPVDFRRV 148
+ +K G+V V+++RV
Sbjct: 428 GKNADASAELFKYGVVDVEYKRV 496
>TC8270 similar to UP|Q7XHJ2 (Q7XHJ2) Expansin-like protein (Fragment),
partial (90%)
Length = 1006
Score = 53.5 bits (127), Expect = 1e-08
Identities = 40/121 (33%), Positives = 57/121 (47%), Gaps = 8/121 (6%)
Frame = +3
Query: 36 SDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIRCANDHRWCLPGS-I 94
S AS GACGY +L G + AA +LF +G CG+CY++RC N G+ +
Sbjct: 129 SKASALSSGACGYDSLALGLSGGHLAAGVPSLFKSGAGCGACYQLRCKNPTLCKKEGARV 308
Query: 95 VVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQ-------YKAGIVPVDFRR 147
V+T N PNN F L+ F+ +AQ K GIV ++++R
Sbjct: 309 VLTDLN--------PNNQ--------TEFVLSSRAFVAMAQDGKGQQILKLGIVDIEYKR 440
Query: 148 V 148
V
Sbjct: 441 V 443
>TC15173 similar to UP|AAR09168 (AAR09168) Alpha-expansin 1, partial (50%)
Length = 898
Score = 47.4 bits (111), Expect = 1e-06
Identities = 20/31 (64%), Positives = 26/31 (83%)
Frame = +2
Query: 118 PPLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
PP H DLA P+FL+IA+Y++GIVPV +RRV
Sbjct: 2 PPRPHSDLAMPMFLKIAEYRSGIVPVAYRRV 94
>TC8271 weakly similar to UP|EXL2_ARATH (Q9SVE5) Expansin-like 2 precursor
(At-EXPL2) (Ath-ExpBeta-2.2), partial (38%)
Length = 465
Score = 40.0 bits (92), Expect = 2e-04
Identities = 19/45 (42%), Positives = 26/45 (57%)
Frame = +1
Query: 36 SDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEI 80
S AS GACGY +L G + AA +LF +G CG+CY++
Sbjct: 139 SKASALSSGACGYDSLALGLSGGHLAAGVPSLFKSGAGCGACYQV 273
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.140 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,390,784
Number of Sequences: 28460
Number of extensions: 56787
Number of successful extensions: 360
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 341
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 346
length of query: 148
length of database: 4,897,600
effective HSP length: 82
effective length of query: 66
effective length of database: 2,563,880
effective search space: 169216080
effective search space used: 169216080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 51 (24.3 bits)
Medicago: description of AC140030.4


