

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140025.4 + phase: 0
(277 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
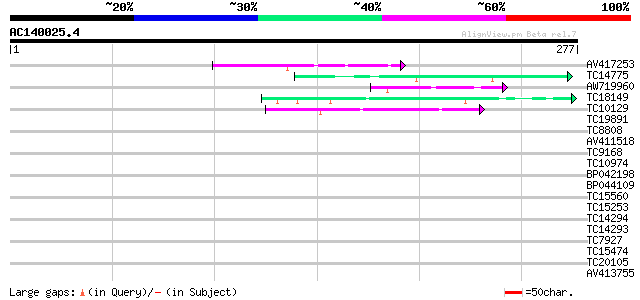
Score E
Sequences producing significant alignments: (bits) Value
AV417253 56 6e-09
TC14775 PIR|S14181|S14181 DNA-directed RNA polymerase largest c... 44 3e-05
AW719960 43 7e-05
TC18149 weakly similar to UP|Q91BL3 (Q91BL3) Essential structura... 42 1e-04
TC10129 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (10%) 40 3e-04
TC19891 homologue to PIR|S14182|S14182 DNA-directed RNA polymera... 39 0.001
TC8808 37 0.003
AV411518 37 0.003
TC9168 similar to GB|AAP21369.1|30102902|BT006561 At4g19200 {Ara... 37 0.004
TC10974 36 0.006
BP042198 35 0.011
BP044109 35 0.011
TC15560 similar to SP|Q03173|NDPP_MOUSE NPC derived proline rich... 35 0.014
TC15253 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-pro... 35 0.018
TC14294 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding pr... 34 0.031
TC14293 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding pr... 34 0.031
TC7927 similar to UP|Q93YR3 (Q93YR3) HSP associated protein like... 33 0.053
TC15474 UP|Q7Y1B9 (Q7Y1B9) Ammonium transporter, complete 32 0.091
TC20105 homologue to GB|AAM61695.1|21537354|AY085142 hydroxyprol... 32 0.15
AV413755 32 0.15
>AV417253
Length = 323
Score = 56.2 bits (134), Expect = 6e-09
Identities = 38/102 (37%), Positives = 48/102 (46%), Gaps = 8/102 (7%)
Frame = +3
Query: 100 KLHLSYSRHTDLNVKAFSDKSRDYTVPLVPAPVWQ--------NPQAAPMYPTNSPAYQT 151
KLH+SYSRHTDL++K +D+SRDYT+P P Q P P N Y T
Sbjct: 6 KLHISYSRHTDLSIKVNNDRSRDYTIPTTPVVNTQPSILGQQPGPLVGPAQQYNGSQY-T 182
Query: 152 QVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQ 193
+ S Q+Q G P+ + YVP PG P Q
Sbjct: 183 PISSQSLTPQSQA-GWGAPAQSMPMQMHNNMYVP-PGTMPPQ 302
>TC14775 PIR|S14181|S14181 DNA-directed RNA polymerase largest chain
(isoform B1) - soybean (fragment) {Glycine
max;} , partial (22%)
Length = 848
Score = 43.9 bits (102), Expect = 3e-05
Identities = 40/139 (28%), Positives = 55/139 (38%), Gaps = 3/139 (2%)
Frame = +1
Query: 140 PMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAF-P 198
P Y SPAY PG SP PS+ T P Y P +Y Q+ + P
Sbjct: 1 PSYSPTSPAYSPTSPGYSPT---------SPSYSPTS----PSYSPTSPSYNPQSAKYSP 141
Query: 199 TMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGG--STSGPGSSPHMQQNLGAQGMVR 256
++ S+ + SSP + +S +P + + P+ S S P SP N G
Sbjct: 142 SLAYSPSSPRLSPSSPYSPTSPNYSPTSPSYSPTSPSYSPSSPTYSPSSPYNSGVSPDYS 321
Query: 257 PGAPPNVRPGGASPSGQHY 275
P +P G SPS Y
Sbjct: 322 PSSPQFSPSTGYSPSQPGY 378
Score = 33.9 bits (76), Expect = 0.031
Identities = 20/69 (28%), Positives = 27/69 (38%)
Frame = +1
Query: 139 APMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAFP 198
+P Y SP+Y P SP+ T P S V P Y P + TG P
Sbjct: 202 SPNYSPTSPSYSPTSPSYSPSSPTYSPSSPYNS------GVSPDYSPSSPQFSPSTGYSP 363
Query: 199 TMPSYGSAA 207
+ P Y ++
Sbjct: 364 SQPGYSPSS 390
>AW719960
Length = 449
Score = 42.7 bits (99), Expect = 7e-05
Identities = 28/72 (38%), Positives = 33/72 (44%), Gaps = 5/72 (6%)
Frame = +3
Query: 177 HAVRPGY-----VPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQP 231
HA PGY P PG YPGQ GA +Y A P A P A + +PGA P
Sbjct: 114 HAAAPGYPAPGQYPPPGGYPGQYGAPGAPGAYPGA--PPAGYPGAPAGYPGAYPGA---P 278
Query: 232 SGGSTSGPGSSP 243
+ G PG +P
Sbjct: 279 AAGYPGAPGGAP 314
Score = 27.3 bits (59), Expect = 2.9
Identities = 26/75 (34%), Positives = 31/75 (40%), Gaps = 8/75 (10%)
Frame = -2
Query: 174 LTQHAVRPGYVPVPGAYPGQTGA--------FPTMPSYGSAAMPTASSPLAQSSHPGAPH 225
L + A PG VP PGA PG G P P+ G+ P +P PGAP+
Sbjct: 352 LPEPAEWPG-VP*PGAPPGAPG*PAAGAPG*APG*PA-GAPG*PAGGAPG*APGAPGAPY 179
Query: 226 NVNLQPSGGSTSGPG 240
P GG G G
Sbjct: 178 WPG*PPGGGYCPGAG 134
>TC18149 weakly similar to UP|Q91BL3 (Q91BL3) Essential structural protein
pp78/81, partial (5%)
Length = 710
Score = 42.0 bits (97), Expect = 1e-04
Identities = 45/166 (27%), Positives = 65/166 (39%), Gaps = 12/166 (7%)
Frame = +3
Query: 124 TVPLVP---APVWQNPQAA--PMYPTNSPAYQTQVPG---GSPAYQTQVPGGQVPSWDLT 175
++P+VP AP PQ P +P + QTQ+P P + V + P+
Sbjct: 102 SIPVVPPPNAPPQLPPQQGLPPSFPPPNQFSQTQIPAVPQRDPYFPPPVQSQETPNQQY- 278
Query: 176 QHAVRPGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHP----GAPHNVNLQP 231
Q + P P PGA P Q P Y A P P+ + P H+V P
Sbjct: 279 QQPLAPQPHPQPGAPPHQQYQQTPHPQYSQPAPPQHQPPIPSINPPQLQSSLGHHVEEPP 458
Query: 232 SGGSTSGPGSSPHMQQNLGAQGMVRPGAPPNVRPGGASPSGQHYYG 277
S + P P+++Q P PP+ G +P Q +YG
Sbjct: 459 YVPSQTYP---PNLRQ--------PPSQPPS---GPPAPPSQQFYG 554
>TC10129 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (10%)
Length = 562
Score = 40.4 bits (93), Expect = 3e-04
Identities = 32/110 (29%), Positives = 45/110 (40%), Gaps = 3/110 (2%)
Frame = +1
Query: 126 PLVPAPVWQNPQAAPMYPTNSPAYQ---TQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPG 182
P P+P+ Q P T SP Q T P +P+ +Q P P + +P
Sbjct: 232 PKTPSPISQPPHVPTPPSTPSPISQPPYTPTPPKTPSPTSQPPHTPTPP-NTPSPISQPP 408
Query: 183 YVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPS 232
Y P P P T P +PS +++ PT S P S P P + P+
Sbjct: 409 YTPTPPKTPSPTNQPPHIPSPPNSSSPT-SQPPNTPSPPKTPSPTSQPPN 555
>TC19891 homologue to PIR|S14182|S14182 DNA-directed RNA polymerase largest
chain (isoform B2) - soybean (fragment)
{Glycine max;} , partial (34%)
Length = 498
Score = 38.9 bits (89), Expect = 0.001
Identities = 29/88 (32%), Positives = 35/88 (38%), Gaps = 6/88 (6%)
Frame = +3
Query: 143 PTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPV-PGAYPGQTGAFPTMP 201
PT+SP Y PG SP+ P PGY P PG P G PT P
Sbjct: 273 PTSSPGYSPSSPGYSPSSPGYSP-------------TSPGYSPTSPGYSPTSPGYSPTSP 413
Query: 202 SY-----GSAAMPTASSPLAQSSHPGAP 224
+Y G + A SP + S P +P
Sbjct: 414 TYSPSSPGYSPTSPAYSPTSPSYSPTSP 497
Score = 28.5 bits (62), Expect = 1.3
Identities = 33/133 (24%), Positives = 45/133 (33%), Gaps = 5/133 (3%)
Frame = +3
Query: 148 AYQTQVPGGSPAYQTQVPGGQVPSWDLTQHA--VRPGYVPVPGAYPGQTGAFPTMPSYGS 205
A + Q+P + G+ P H + P Y+ P T P G
Sbjct: 81 AIELQLPSYMDGLDFGMTPGRSPISGTPYHEGMMSPSYMLSPNLRLSPTNDAQFSPYVGG 260
Query: 206 AAMPTASSPLAQSSHPG-APHNVNLQPS--GGSTSGPGSSPHMQQNLGAQGMVRPGAPPN 262
A SSP S PG +P + P+ G S + PG SP P +P
Sbjct: 261 MAFSPTSSPGYSPSSPGYSPSSPGYSPTSPGYSPTSPGYSPTSPGYSPTSPTYSPSSP-- 434
Query: 263 VRPGGASPSGQHY 275
G SP+ Y
Sbjct: 435 ----GYSPTSPAY 461
>TC8808
Length = 579
Score = 37.4 bits (85), Expect = 0.003
Identities = 37/122 (30%), Positives = 47/122 (38%), Gaps = 12/122 (9%)
Frame = +1
Query: 133 WQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPG 192
+ +P AA + P Y PGG P Y Q + +A G P YP
Sbjct: 4 YNHPPAA-----SGPVYAQ--PGGQPTYS------QPSAQPAASYAQGYGSYPSQQTYPE 144
Query: 193 QTGA--------FPTMPSYGSAAMPTASSPLAQSSHPG----APHNVNLQPSGGSTSGPG 240
Q+ P PSYGSAA P S+ AQS PG P + S G + P
Sbjct: 145 QSAPNNAVYGYQAPQDPSYGSAAAPAYSA--AQSGQPGYAQPTPTQTGYEQSAGYAAVPA 318
Query: 241 SS 242
+S
Sbjct: 319 AS 324
>AV411518
Length = 360
Score = 37.4 bits (85), Expect = 0.003
Identities = 30/88 (34%), Positives = 40/88 (45%), Gaps = 1/88 (1%)
Frame = +3
Query: 185 PVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGS-SP 243
P P A+P A + S+ S PT+S P + S AP + +P+ STSGP S +P
Sbjct: 93 PSP*AHPTPAWAGSALASWASPCAPTSSGPASPSPSSTAP---SPRPNPSSTSGPTSPTP 263
Query: 244 HMQQNLGAQGMVRPGAPPNVRPGGASPS 271
H P PP P +SPS
Sbjct: 264 H-----------TPSPPP---PTSSSPS 305
>TC9168 similar to GB|AAP21369.1|30102902|BT006561 At4g19200 {Arabidopsis
thaliana;}, partial (12%)
Length = 682
Score = 37.0 bits (84), Expect = 0.004
Identities = 45/161 (27%), Positives = 56/161 (33%), Gaps = 34/161 (21%)
Frame = +2
Query: 136 PQAAPMY-----PTNSPAY------QTQVPGGSPAYQTQVPGGQVPSWDLTQH------- 177
PQ AP PT+ AY Q+ G Q Q P G P + +Q
Sbjct: 23 PQGAPQSGYGVPPTSQAAYGNPPQVQSGYGAGYGPPQAQKPSGTPPVYGQSQSPNTTGGY 202
Query: 178 ---------------AVRPGY-VPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHP 221
AV+PGY P GA GQ+G P+YGS+A P P
Sbjct: 203 GQTGGYPPSHPPYGGAVQPGYGAPPYGAPGGQSGYAQAPPTYGSSAYGAGYPP------P 364
Query: 222 GAPHNVNLQPSGGSTSGPGSSPHMQQNLGAQGMVRPGAPPN 262
A GG+T G Q QG V +PP+
Sbjct: 365 SAYSGDGNAGEGGNTRGSNDGAPAQ--AVQQGSVAKSSPPS 481
>TC10974
Length = 652
Score = 36.2 bits (82), Expect = 0.006
Identities = 33/102 (32%), Positives = 41/102 (39%), Gaps = 17/102 (16%)
Frame = +3
Query: 184 VPVPGAYPGQTGAF-------PTMPSYGSAAM----PTASSPLA----QSSHPGAPHNVN 228
+P+PG Q GA P P G M P+ PL + P N N
Sbjct: 33 MPMPGLMGMQQGAMNQMGNPMPQGPFVGMNQMHSGPPSGGPPLGGFPNHLPNMQGPSNTN 212
Query: 229 LQPSGGSTSGP--GSSPHMQQNLGAQGMVRPGAPPNVRPGGA 268
P G S + P G P MQ Q +PG PPN +PGG+
Sbjct: 213 Y-PQGASFNRPQGGQMPMMQGYNPYQSGNQPGMPPNAQPGGS 335
>BP042198
Length = 481
Score = 35.4 bits (80), Expect = 0.011
Identities = 30/119 (25%), Positives = 47/119 (39%), Gaps = 6/119 (5%)
Frame = -3
Query: 143 PTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAFPTMPS 202
P +SP+ P P+ T P + P AV P PV + + + P
Sbjct: 467 PASSPSNNVSSPPPVPSPATPPPTAETP-------AVSPVSSPVAAPTVAKGPSASSSPP 309
Query: 203 YGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSP------HMQQNLGAQGMV 255
SA +PT+S+ A + P + + ++S P SSP N GA G++
Sbjct: 308 EASAGVPTSSATPADAPVTTIPSSSTAPGTAPASSSPESSPGAADDSGSSSNFGAPGLL 132
>BP044109
Length = 497
Score = 35.4 bits (80), Expect = 0.011
Identities = 37/143 (25%), Positives = 57/143 (38%), Gaps = 3/143 (2%)
Frame = +2
Query: 103 LSYSRHTDLNVKAFSDKSRDYTVPLVPAPVWQNPQAAPMYPTNSPAY---QTQVPGGSPA 159
LS+S T L+ F + P P P + P + +N+ + +TQ P P
Sbjct: 59 LSHSSSTSLSSSHF------FPSPPPPPPPPNKNKTTPSHASNATSKSTPRTQTPTTPPP 220
Query: 160 YQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSS 219
T P PS T+ + P P ++ G PT+PS P++S+P + S
Sbjct: 221 SPTSSPKPP-PSTFATKPSTSPPETPSSSSH----GPAPTLPS-----PPSSSTPTSTPS 370
Query: 220 HPGAPHNVNLQPSGGSTSGPGSS 242
P P+ L +T P S
Sbjct: 371 PPSPPNGSTLPSPPRATPTPAPS 439
>TC15560 similar to SP|Q03173|NDPP_MOUSE NPC derived proline rich protein 1
(NDPP-1). {Mus musculus;}, partial (8%)
Length = 1134
Score = 35.0 bits (79), Expect = 0.014
Identities = 29/116 (25%), Positives = 45/116 (38%)
Frame = +3
Query: 129 PAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPG 188
P+P P P+ P +P + +P P+ T P P +P P+P
Sbjct: 171 PSPSLPPPSPPPLPPPQAPPLPSHLPSPEPS-STMPPSTPPPQ------PTKP--CPLPK 323
Query: 189 AYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPH 244
+ P Q PS +PT+SS + +S PH+ + ST SP+
Sbjct: 324 STPSQP------PSSAPHRVPTSSSLASPTSPSSGPHSTTMAAPFSSTRTSTPSPN 473
>TC15253 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-protein
(ATAGP4) (AT5G10430/F12B17_220), partial (42%)
Length = 575
Score = 34.7 bits (78), Expect = 0.018
Identities = 33/109 (30%), Positives = 37/109 (33%)
Frame = +3
Query: 152 QVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAFPTMPSYGSAAMPTA 211
Q PG +P TQ P P A P P P A P T P SA+ P
Sbjct: 147 QAPGAAP---TQAPTTTPPPPPAAAPAPPPA-TPPPAATPAPTTTPPAATPAPSASPP-- 308
Query: 212 SSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPHMQQNLGAQGMVRPGAP 260
+P +S GAP P SGP S P G P P
Sbjct: 309 -APTPTASPTGAPTPSASSPPAPIPSGPASGPGPAAGPGPNSADTPPPP 452
>TC14294 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding protein-like
(At5g04170), partial (65%)
Length = 1192
Score = 33.9 bits (76), Expect = 0.031
Identities = 35/103 (33%), Positives = 45/103 (42%), Gaps = 3/103 (2%)
Frame = +1
Query: 174 LTQHAVRPGYVPVPGAYPGQTGAFPTMPSYGSA--AMPTASSPLAQSSHPGAPHNVNLQP 231
+ H GY P Y GA P YG+A + P ++P +QS GAP QP
Sbjct: 43 IPSHFNMSGYPNKPSGYG--YGAPPPYQPYGAAPPSQPYGAAPPSQSY--GAPPPP--QP 204
Query: 232 SGGST-SGPGSSPHMQQNLGAQGMVRPGAPPNVRPGGASPSGQ 273
G + S P +P Q+ G G PP +P ASP GQ
Sbjct: 205 YGAAPPSQPYGAPPPSQSYG-------GPPPPSQPYSASPYGQ 312
Score = 32.7 bits (73), Expect = 0.069
Identities = 42/142 (29%), Positives = 51/142 (35%), Gaps = 12/142 (8%)
Frame = +1
Query: 142 YPTNSPAYQTQVPGGSPAYQ-------TQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQT 194
YP Y P P YQ +Q G PS + P P A P Q
Sbjct: 70 YPNKPSGYGYGAP---PPYQPYGAAPPSQPYGAAPPS---QSYGAPPPPQPYGAAPPSQP 231
Query: 195 -GAFPTMPSYGS---AAMPTASSPLAQSSHP-GAPHNVNLQPSGGSTSGPGSSPHMQQNL 249
GA P SYG + P ++SP Q S P AP+ + S G G S +
Sbjct: 232 YGAPPPSQSYGGPPPPSQPYSASPYGQPSAPYAAPYQKPPKDESHSHGGGGGSGYPPPP- 408
Query: 250 GAQGMVRPGAPPNVRPGGASPS 271
A G P+V P G PS
Sbjct: 409 SAYGSPFASLVPSVFPPGTDPS 474
>TC14293 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding protein-like
(At5g04170), partial (18%)
Length = 540
Score = 33.9 bits (76), Expect = 0.031
Identities = 41/135 (30%), Positives = 49/135 (35%), Gaps = 5/135 (3%)
Frame = +2
Query: 142 YPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQT-GAFPTM 200
YP Y P P YQ P G P + P P A P Q GA P
Sbjct: 110 YPNKPSGYGYGAP---PPYQ---PYGAAPPSQ--SYGAPPPPQPYGAAPPSQPYGAPPPS 265
Query: 201 PSYGS---AAMPTASSPLAQSSHP-GAPHNVNLQPSGGSTSGPGSSPHMQQNLGAQGMVR 256
SYG + P ++SP Q S P AP+ + S G G S + A G
Sbjct: 266 QSYGGPPPPSQPYSASPYGQPSAPYAAPYQKPPKDESHSHGGGGGSGYPPPP-SAYGSPF 442
Query: 257 PGAPPNVRPGGASPS 271
P+V P G PS
Sbjct: 443 ASLVPSVFPPGTDPS 487
Score = 28.9 bits (63), Expect = 1.0
Identities = 30/83 (36%), Positives = 38/83 (45%), Gaps = 6/83 (7%)
Frame = +2
Query: 197 FPTMPS-YGSAA----MPTASSPLAQSSHPGAPHNVNLQPSGGST-SGPGSSPHMQQNLG 250
+P PS YG A P ++P +QS GAP QP G + S P +P Q+ G
Sbjct: 110 YPNKPSGYGYGAPPPYQPYGAAPPSQSY--GAPPPP--QPYGAAPPSQPYGAPPPSQSYG 277
Query: 251 AQGMVRPGAPPNVRPGGASPSGQ 273
G PP +P ASP GQ
Sbjct: 278 -------GPPPPSQPYSASPYGQ 325
>TC7927 similar to UP|Q93YR3 (Q93YR3) HSP associated protein like, partial
(56%)
Length = 1116
Score = 33.1 bits (74), Expect = 0.053
Identities = 21/59 (35%), Positives = 27/59 (45%), Gaps = 3/59 (5%)
Frame = -2
Query: 215 LAQSSHPGAPHNVNLQPSGGSTSG---PGSSPHMQQNLGAQGMVRPGAPPNVRPGGASP 270
L +S+ PG P + P G+ G PG+ P M GM PG PP + P G P
Sbjct: 674 LLKSTFPGTPPGM---PPPGNPPGMPPPGNPPGMPPPGNPPGMPPPGKPPGMPPPGNPP 507
Score = 30.0 bits (66), Expect = 0.45
Identities = 26/77 (33%), Positives = 30/77 (38%), Gaps = 1/77 (1%)
Frame = -2
Query: 195 GAFPTMPSYGSA-AMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPHMQQNLGAQG 253
G P MP G+ MP +P PG P N P G PG P M G
Sbjct: 653 GTPPGMPPPGNPPGMPPPGNP------PGMPPPGN--PPG--MPPPGKPPGMPPPGNPPG 504
Query: 254 MVRPGAPPNVRPGGASP 270
+ PG PP + P G P
Sbjct: 503 IPPPGNPPGIPPPGNPP 453
Score = 27.7 bits (60), Expect = 2.2
Identities = 28/91 (30%), Positives = 31/91 (33%), Gaps = 4/91 (4%)
Frame = +1
Query: 186 VPGAYPGQTGAFP--TMP-SYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSG-PGS 241
+PG +PG G FP MP + MP PG P GG G PG
Sbjct: 427 MPGEFPGMPGGFPGGGMPGGFPGGGMP---GGFPGGGMPGG------FPGGGMPGGFPGG 579
Query: 242 SPHMQQNLGAQGMVRPGAPPNVRPGGASPSG 272
G G G P PGG P G
Sbjct: 580 --------GMPGGFPGGGMPGGFPGGGMPGG 648
>TC15474 UP|Q7Y1B9 (Q7Y1B9) Ammonium transporter, complete
Length = 1950
Score = 32.3 bits (72), Expect = 0.091
Identities = 31/110 (28%), Positives = 42/110 (38%), Gaps = 4/110 (3%)
Frame = +3
Query: 138 AAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAF 197
A P P +P +Q+ SP PS T P P Q A+
Sbjct: 180 ATPPMPLTTPTSSSQLI*SSPCSSVLPCSAPAPSEPKT-----P*TSC*PMFLMQQPAAY 344
Query: 198 PTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQP----SGGSTSGPGSSP 243
PT S ++ P+ P A S+ + N L+P + STSGP SP
Sbjct: 345 PTTSS--ASRSPSEPPPTASSAATSSASNTTLRPPTTTASSSTSGPSPSP 488
>TC20105 homologue to GB|AAM61695.1|21537354|AY085142 hydroxyproline-rich
glycoprotein-like protein {Arabidopsis thaliana;},
partial (7%)
Length = 603
Score = 31.6 bits (70), Expect = 0.15
Identities = 25/86 (29%), Positives = 35/86 (40%), Gaps = 2/86 (2%)
Frame = +3
Query: 127 LVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQT--QVPGGQVPSWDLTQHAVRPGYV 184
L A V P AAP SP++ Q P P Y + + G + P + A P
Sbjct: 69 LTNAYVSSFPAAAPPTFVRSPSHG-QYPAALPPYPSPPHMYGSRSPPY-----AYSPEPA 230
Query: 185 PVPGAYPGQTGAFPTMPSYGSAAMPT 210
P+ +YP +P YG+ PT
Sbjct: 231 PIAASYPAAPMNYPAYGGYGNVLAPT 308
>AV413755
Length = 424
Score = 31.6 bits (70), Expect = 0.15
Identities = 27/112 (24%), Positives = 41/112 (36%)
Frame = +3
Query: 129 PAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPG 188
P P NPQ P SP+ P +P + P G P ++ H G P P
Sbjct: 93 PYPGSTNPQMTP-----SPSQVFPAPYRNPVWNG--PRGPAP-YNFPFHPSGGGTYPNPR 248
Query: 189 AYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPG 240
P + ++ PSY P ++ + P + N +P+ PG
Sbjct: 249 FEPSSSPSYNYRPSYSPNPSPGYTNTPSPRFEPSRSPSYNYRPNYSPNPSPG 404
Score = 26.2 bits (56), Expect = 6.5
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 10/40 (25%)
Frame = +2
Query: 190 YPGQTGAFPTMP----SYGSAAM------PTASSPLAQSS 219
YPG TG FP+ SYG+ ++ +A PL+QSS
Sbjct: 23 YPGLTGLFPSSKYWWFSYGTISITISRINKSADDPLSQSS 142
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.130 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,807,829
Number of Sequences: 28460
Number of extensions: 72183
Number of successful extensions: 517
Number of sequences better than 10.0: 103
Number of HSP's better than 10.0 without gapping: 482
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 504
length of query: 277
length of database: 4,897,600
effective HSP length: 89
effective length of query: 188
effective length of database: 2,364,660
effective search space: 444556080
effective search space used: 444556080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC140025.4


