

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140022.4 - phase: 0
(205 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
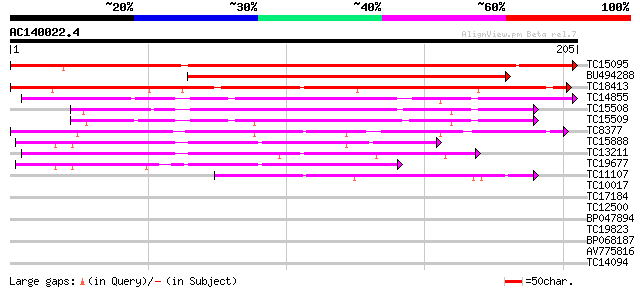
Score E
Sequences producing significant alignments: (bits) Value
TC15095 similar to UP|AAQ96377 (AAQ96377) Miraculin-like protein... 311 5e-86
BU494288 187 1e-48
TC18413 similar to UP|P93378 (P93378) Tumor-related protein, par... 165 5e-42
TC14855 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precurs... 103 2e-23
TC15508 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precurs... 92 5e-20
TC15509 weakly similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor ... 86 3e-18
TC8377 similar to UP|Q9M3Z7 (Q9M3Z7) Alpha-fucosidase , partial... 82 5e-17
TC15888 weakly similar to UP|Q8LK19 (Q8LK19) Proteinase inhibito... 74 2e-14
TC13211 weakly similar to UP|Q8LK19 (Q8LK19) Proteinase inhibito... 69 6e-13
TC19677 weakly similar to UP|O82711 (O82711) Profucosidase precu... 57 2e-09
TC11107 44 3e-05
TC10017 UP|RR3_LOTJA (Q9BBP8) Chloroplast 30S ribosomal protein ... 27 1.9
TC17184 similar to UP|Q8S9J9 (Q8S9J9) At1g14000/F7A19_9, partial... 27 2.5
TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein... 27 2.5
BP047894 27 3.3
TC19823 27 3.3
BP068187 26 5.6
AV775816 26 5.6
TC14094 homologue to emb|X02559.1|MIOBRN26 Oenothera berteriana ... 25 7.3
>TC15095 similar to UP|AAQ96377 (AAQ96377) Miraculin-like protein
(Fragment), partial (42%)
Length = 979
Score = 311 bits (797), Expect = 5e-86
Identities = 158/211 (74%), Positives = 177/211 (83%), Gaps = 6/211 (2%)
Frame = +3
Query: 1 MKTSFLAFSIIFLAFICK------TFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLT 54
MKTS LAFSII AF + + AAPEPVLDISG+K+ TGVKYYILPV+RGKGGGLT
Sbjct: 186 MKTSQLAFSIICFAFTTEFLIGIASAAAPEPVLDISGQKLRTGVKYYILPVLRGKGGGLT 365
Query: 55 VVNENNLNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTC 114
+ + N+N NNTCPL+V+QEKLEV GQ+VTFTPYNAK GVILTSTDLNIKS +T TTC
Sbjct: 366 LTSSGNIN--NNTCPLHVVQEKLEVLKGQSVTFTPYNAKGGVILTSTDLNIKSSLTNTTC 539
Query: 115 AQSQVWKLNKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVCKCQTLC 174
A++ VWKL K LSGVWFLATGGVEGNPG TI NWFKIEKADKDYVFSFCPSVCKCQTLC
Sbjct: 540 AKAPVWKLLKELSGVWFLATGGVEGNPGMATISNWFKIEKADKDYVFSFCPSVCKCQTLC 719
Query: 175 RELGLYVYDHGKKHLALSDQVPSFRVVFKRA 205
RELG+Y Y G KHL+LSDQVP FR++FK+A
Sbjct: 720 RELGIYDYG-GDKHLSLSDQVPPFRIMFKQA 809
>BU494288
Length = 438
Score = 187 bits (475), Expect = 1e-48
Identities = 88/117 (75%), Positives = 95/117 (80%)
Frame = +3
Query: 65 NNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKLNK 124
N +CP V+Q KLEV NG VTFTPYNAK GVILTSTDLNIKS+ K+ C S VWKL K
Sbjct: 30 NKSCPPNVVQAKLEVWNGTPVTFTPYNAKDGVILTSTDLNIKSFPIKSPCGPSFVWKLQK 209
Query: 125 VLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVCKCQTLCRELGLYV 181
L+GVWFL TGGVEGNPG DTI NWFKIEKA KDYV SFCPSVCKC TLCRELG+Y+
Sbjct: 210 ELTGVWFLVTGGVEGNPGADTIVNWFKIEKAGKDYVLSFCPSVCKCNTLCRELGIYI 380
Score = 26.2 bits (56), Expect = 4.3
Identities = 16/49 (32%), Positives = 22/49 (44%)
Frame = +1
Query: 151 KIEKADKDYVFSFCPSVCKCQTLCRELGLYVYDHGKKHLALSDQVPSFR 199
++ K +K F P LG + HLALSD+VPSF+
Sbjct: 289 RLRKLEKTMCFLSVPQSANAILYAGNLGSILMVR-TSHLALSDKVPSFK 432
>TC18413 similar to UP|P93378 (P93378) Tumor-related protein, partial (41%)
Length = 740
Score = 165 bits (418), Expect = 5e-42
Identities = 99/215 (46%), Positives = 134/215 (62%), Gaps = 12/215 (5%)
Frame = -3
Query: 1 MKTSFLAFSIIFLA----FICKTFAAPEPVLDISGKKVTTGVKYYILPVIRGK--GGGLT 54
MK+ +L+F+I+F F+ K A+PE V+D GK V G+ YYI PV G G
Sbjct: 714 MKSIYLSFAILFALCTQPFLGKGDASPEQVVDTQGKNVRAGMGYYIRPVPTTPCDGRGPC 535
Query: 55 VVNENNL---NGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTK 111
VV + N TCPL V+ +E GQAVTFTP N KKGVI STD+NI + +
Sbjct: 534 VVGSGFVLIARSPNKTCPLNVVV--VEGFRGQAVTFTPVNPKKGVIRVSTDVNI-NVTLE 364
Query: 112 TTCAQSQVWKLNK--VLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVC- 168
T C +S VWKL+ +G WF+ TGGV GNPG DTI NWFKIEK ++DY FCP+VC
Sbjct: 363 TACEESTVWKLDDFDASAGHWFVTTGGVVGNPGKDTISNWFKIEKYEEDYKLVFCPTVCD 184
Query: 169 KCQTLCRELGLYVYDHGKKHLALSDQVPSFRVVFK 203
C+ LCR +G+++ +G + +AL+D+ S++V F+
Sbjct: 183 TCKPLCRNVGVFMDSNGNRRVALTDE--SYKVRFQ 85
>TC14855 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precursor, partial
(90%)
Length = 1001
Score = 103 bits (257), Expect = 2e-23
Identities = 70/206 (33%), Positives = 107/206 (50%), Gaps = 5/206 (2%)
Frame = +1
Query: 5 FLAFSIIFLAFICKTFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGN 64
FL+ I+ + PEPV+D G + GV YY P + GGLT+ N
Sbjct: 76 FLSLLILNTQPLLAAEPEPEPVVDKQGNPLEPGVGYYAWP-LWADNGGLTLGRTRN---- 240
Query: 65 NNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKLNK 124
TCPL ++++ ++ G + F ++ G I T TDL ++ + + C + +VW+L K
Sbjct: 241 -KTCPLDIIRDPSKI--GTPIEFHVADSDLGYIPTLTDLTVEMPILGSRCQEPKVWRLLK 411
Query: 125 VLSGVWFLATGGVEGNPGFDTIFNWFKIEK-----ADKDYVFSFCPSVCKCQTLCRELGL 179
V SG WFL+TGGV GN + + FKIE+ A + Y F FCPSV LC +G
Sbjct: 412 VGSGFWFLSTGGVAGN-----LVSKFKIERLAGEHAYEIYSFKFCPSV--PGVLCAPVGT 570
Query: 180 YVYDHGKKHLALSDQVPSFRVVFKRA 205
Y G + +A+ D + ++ V F++A
Sbjct: 571 YEDADGTQVMAIGDDIETYYVRFQKA 648
>TC15508 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precursor, partial
(66%)
Length = 877
Score = 92.4 bits (228), Expect = 5e-20
Identities = 61/176 (34%), Positives = 92/176 (51%), Gaps = 7/176 (3%)
Frame = +3
Query: 23 PEP-VLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKN 81
PEP V+D+ G + GV+YY+ PV KGG L + + N TCPL ++ + +
Sbjct: 108 PEPAVVDMQGNPLLPGVQYYVRPVSAEKGG-LALGHTRN-----KTCPLDIILDPSSIIG 269
Query: 82 GQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKLNKVLSGVWFLATGGVEGNP 141
V T ++ G I STDL ++ + + C + +VW+L KV S WF++T GV GN
Sbjct: 270 SPTVFHTSSSSDVGYIPISTDLTVEMPILGSPCKEPKVWRLAKVGSDFWFVSTQGVPGN- 446
Query: 142 GFDTIFNWFKIEKADKD------YVFSFCPSVCKCQTLCRELGLYVYDHGKKHLAL 191
+ F + E+ D Y FSFCPSV +C +G+YV D G + +A+
Sbjct: 447 -LVSKFKIVRFEERDNHANEGEIYSFSFCPSV--YGVICAPVGIYVDDDGTQVMAV 605
>TC15509 weakly similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precursor,
partial (77%)
Length = 797
Score = 86.3 bits (212), Expect = 3e-18
Identities = 58/177 (32%), Positives = 92/177 (51%), Gaps = 8/177 (4%)
Frame = +1
Query: 23 PEPV-LDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKN 81
PEP +D+ G + GVKYY+ PV + GGL + + N TCPL ++ + +
Sbjct: 85 PEPAAVDMQGNPILPGVKYYVRPV-SAENGGLALGHTRN-----KTCPLDIILDPFSL-- 240
Query: 82 GQAVTF-TPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKLNKVLSGVWFLATGGVEGN 140
G F T ++ G I STDL ++ V K+ C + +VW++ K + WF++T GV GN
Sbjct: 241 GTPTVFQTSSSSDLGYIPISTDLTVEMPVLKSPCKEPKVWRIAKEGADFWFVSTQGVPGN 420
Query: 141 PGFDTIFNWFKIEKADKD------YVFSFCPSVCKCQTLCRELGLYVYDHGKKHLAL 191
+ F + E+ D Y FS+CPS+ +C +G+YV D G + +A+
Sbjct: 421 DA--SRFKIVRFEEGDNHANEGEIYSFSYCPSI--WGYICAPVGIYVDDDGTQVMAV 579
>TC8377 similar to UP|Q9M3Z7 (Q9M3Z7) Alpha-fucosidase , partial (83%)
Length = 1014
Score = 82.4 bits (202), Expect = 5e-17
Identities = 68/217 (31%), Positives = 98/217 (44%), Gaps = 15/217 (6%)
Frame = +1
Query: 1 MKTSFLAFSIIFLAFICKTFAAP------EPVLDISGKKVTTGVKYYILPVIRGKGGGLT 54
MK L+ +F AFI E VLD++G + G +YYI P IRG GG
Sbjct: 31 MKPIPLSLCFLFFAFISTKLPTAFSSNDVEQVLDVNGDSIFPGGRYYIRPAIRGPPGGGV 210
Query: 55 VVNENNLNGNNNTCPLYVLQEKLEVKNGQAVTF-TPYNAKKGVILTSTDLNIKSYVTKTT 113
+ + ++ CP+ LQE EVKNG V F G+I T T+L+I+ + K
Sbjct: 211 RLGKT----GDSDCPVTALQEYSEVKNGIPVRFRIAAEVSTGIIFTGTELDIE-FTVKPH 375
Query: 114 CAQSQVW------KLNKVLSGVWFLATGGVEGNPGFDTIFNWFKIEK--ADKDYVFSFCP 165
C +S W ++ K G+ GG + +PG T F I+K A Y FC
Sbjct: 376 CVESPKWIVFVDNEIGKACVGI-----GGAKDHPGKQTFSGTFNIQKNSAGFGYKLVFC- 537
Query: 166 SVCKCQTLCRELGLYVYDHGKKHLALSDQVPSFRVVF 202
K C ++G Y G + L L++ +F +VF
Sbjct: 538 --IKGSPTCLDIGRYDNGEGGRRLNLTEH-EAFDLVF 639
>TC15888 weakly similar to UP|Q8LK19 (Q8LK19) Proteinase inhibitor 20,
partial (27%)
Length = 611
Score = 73.9 bits (180), Expect = 2e-14
Identities = 55/160 (34%), Positives = 74/160 (45%), Gaps = 6/160 (3%)
Frame = +3
Query: 3 TSFLAFSIIFLAF----ICKTFA-APEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVN 57
T L S++ AF I F+ P+ V D +G + + +YYI P +RG GGG
Sbjct: 54 TMLLTLSVLLFAFTTYIIPSAFSQVPDLVKDTNGIPLRSDGRYYIRPALRGPGGGGVSPG 233
Query: 58 ENNLNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQS 117
E N TCP+ VLQ K EV+NG V F Y G+I L I CA S
Sbjct: 234 ETG----NQTCPVTVLQHKSEVENGNGVKFEHY-PNIGIIYEGVPLTISFGDIYGFCADS 398
Query: 118 QVW-KLNKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKAD 156
W ++ G W + GG E +PG + + F I+K D
Sbjct: 399 TKWVVVSDDFPGKW-VGIGGAEDHPGKEIVSGLFDIQKYD 515
>TC13211 weakly similar to UP|Q8LK19 (Q8LK19) Proteinase inhibitor 20,
partial (44%)
Length = 558
Score = 68.9 bits (167), Expect = 6e-13
Identities = 54/171 (31%), Positives = 79/171 (45%), Gaps = 5/171 (2%)
Frame = +1
Query: 5 FLAFSIIFLAFICKTFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGN 64
F F++ F PE V D G + G +Y I+P +RG G ++
Sbjct: 49 FFLFALTPYLFPLAFSQDPEQVKDAYGNPLVPGRRYSIVPALRGSRSGGVSPDKTG---- 216
Query: 65 NNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGV--ILTSTDLNIKSYVTKTTCAQSQVWKL 122
N TCPL V+QE E ++G+ V F+ G ILT T L+I ++V K CA + W +
Sbjct: 217 NQTCPLTVVQEYFECRDGKPVIFSVSGITPGTGTILTGTHLDI-AFVEKPECADTSKWVV 393
Query: 123 NKVLSGVWF--LATGGVEGNPGFDTIFNWFKIEKAD-KDYVFSFCPSVCKC 170
L+ ++ GGVE +PG + F I+K D Y FC +C
Sbjct: 394 VGNLTDYPGAPVSIGGVEDHPGQHILDGTFNIQKLDCGGYKLVFCSIENRC 546
>TC19677 weakly similar to UP|O82711 (O82711) Profucosidase precursor ,
partial (27%)
Length = 471
Score = 57.0 bits (136), Expect = 2e-09
Identities = 48/147 (32%), Positives = 70/147 (46%), Gaps = 7/147 (4%)
Frame = +3
Query: 3 TSFLAFSIIFLAF-----ICKTFA-APEPVLDISGKKVTTGVKYYILPVIRG-KGGGLTV 55
T L S++ AF I F+ P+ V DI+G +++ KYY+LP I G GGG++
Sbjct: 45 TILLTLSVLLFAFSTTYIIPLAFSQVPDLVKDINGIPLSSDGKYYVLPAIWGPTGGGVS- 221
Query: 56 VNENNLNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCA 115
+ G N TCP+ VLQ EV+NG+ V F + G+I L I CA
Sbjct: 222 ---PRVTG-NQTCPVTVLQSYYEVENGKGVKFEHF-PNIGIIYEGVPLKIYFANIYGYCA 386
Query: 116 QSQVWKLNKVLSGVWFLATGGVEGNPG 142
+S W + ++ GG +PG
Sbjct: 387 ESTEWVVVSDDFPTPWVGIGGAADHPG 467
>TC11107
Length = 841
Score = 43.5 bits (101), Expect = 3e-05
Identities = 34/121 (28%), Positives = 53/121 (43%), Gaps = 4/121 (3%)
Frame = +3
Query: 75 EKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKLN--KVLSGVWFL 132
E ++ G V FTP+ + D + + TTC QS W + + SG +
Sbjct: 3 ENTDLPEGFPVKFTPFAKGDKYVKLGKDFTVV-FSAATTCVQSTAWSVGSTEAKSGRRLV 179
Query: 133 ATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPS-VCK-CQTLCRELGLYVYDHGKKHLA 190
TGG G + + F +E Y +CP+ VC C+ C G+ + ++GK LA
Sbjct: 180 VTGGDVGERAYGSYFRIVAVEGRAGIYNIQWCPTDVCNICRFRCGTAGI-LRENGKILLA 356
Query: 191 L 191
L
Sbjct: 357 L 359
>TC10017 UP|RR3_LOTJA (Q9BBP8) Chloroplast 30S ribosomal protein S3,
complete
Length = 1185
Score = 27.3 bits (59), Expect = 1.9
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Frame = -1
Query: 133 ATGGVEGNPGFDTIF---NWFKIEKADKDYVFSFCP 165
++ G+ GNP F F N+F ++ + Y+ SFCP
Sbjct: 573 SSNGL*GNPTFSDPFYACNFFSVDTSRNLYLNSFCP 466
>TC17184 similar to UP|Q8S9J9 (Q8S9J9) At1g14000/F7A19_9, partial (33%)
Length = 598
Score = 26.9 bits (58), Expect = 2.5
Identities = 15/48 (31%), Positives = 29/48 (60%), Gaps = 1/48 (2%)
Frame = -1
Query: 29 ISGKKVTTGVKYYILPVIRG-KGGGLTVVNENNLNGNNNTCPLYVLQE 75
I+GK+V+ K+YI+ +RG + G +N + ++G +N +Y L +
Sbjct: 568 INGKRVSMENKFYIMDDLRGMEERGDIFLNLSGISGYSNAPGIYELNK 425
>TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein {Danio
rerio;} , partial (5%)
Length = 678
Score = 26.9 bits (58), Expect = 2.5
Identities = 12/38 (31%), Positives = 19/38 (49%)
Frame = +3
Query: 90 YNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKLNKVLS 127
YN+ + + + N S+ T TT + QVW+ N S
Sbjct: 435 YNSISSKLTHNNNPNSTSFTTTTTISSPQVWRCNHTSS 548
>BP047894
Length = 533
Score = 26.6 bits (57), Expect = 3.3
Identities = 10/24 (41%), Positives = 13/24 (53%)
Frame = +2
Query: 165 PSVCKCQTLCRELGLYVYDHGKKH 188
P CKCQ + E+ Y+ DH H
Sbjct: 149 PRKCKCQVITIEILAYIADHSILH 220
>TC19823
Length = 452
Score = 26.6 bits (57), Expect = 3.3
Identities = 15/37 (40%), Positives = 19/37 (50%)
Frame = -2
Query: 80 KNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQ 116
K G ++T +PY A G TS DLN + TC Q
Sbjct: 184 KVGFSLTISPY*AFSGTSSTSKDLNHEF*ANYQTCLQ 74
>BP068187
Length = 509
Score = 25.8 bits (55), Expect = 5.6
Identities = 15/25 (60%), Positives = 17/25 (68%), Gaps = 3/25 (12%)
Frame = -2
Query: 165 PSVCKCQTL-C--RELGLYVYDHGK 186
PS+C CQTL C R L L+V D GK
Sbjct: 229 PSLCLCQTLPCT*RLLFLFVLDFGK 155
>AV775816
Length = 389
Score = 25.8 bits (55), Expect = 5.6
Identities = 15/52 (28%), Positives = 25/52 (47%), Gaps = 8/52 (15%)
Frame = +1
Query: 111 KTTCAQSQVWKLNKVLSGVWFLATGG--------VEGNPGFDTIFNWFKIEK 154
K ++S+VW + K+L G W + GG + NPG+ W++ K
Sbjct: 238 KAQGSKSRVWVIFKLLRGFW*PSRGGTGCSWVVVTKANPGW-----WYQYSK 378
>TC14094 homologue to emb|X02559.1|MIOBRN26 Oenothera berteriana mitochondrial
gene for 26S ribosomal RNA, partial (83%)
Length = 3140
Score = 25.4 bits (54), Expect = 7.3
Identities = 12/33 (36%), Positives = 20/33 (60%), Gaps = 1/33 (3%)
Frame = -2
Query: 89 PYNAKKGVILTST-DLNIKSYVTKTTCAQSQVW 120
P++ GVIL S+ D+ + S + +TC+ VW
Sbjct: 1996 PFSRSYGVILPSSFDMVLSSALVYSTCSPVSVW 1898
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,896,457
Number of Sequences: 28460
Number of extensions: 55890
Number of successful extensions: 317
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 297
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 301
length of query: 205
length of database: 4,897,600
effective HSP length: 86
effective length of query: 119
effective length of database: 2,450,040
effective search space: 291554760
effective search space used: 291554760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 53 (25.0 bits)
Medicago: description of AC140022.4


