

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140022.13 + phase: 0
(213 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
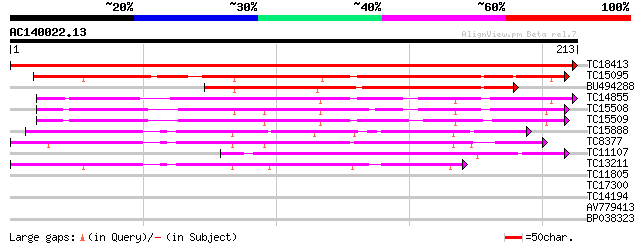
Score E
Sequences producing significant alignments: (bits) Value
TC18413 similar to UP|P93378 (P93378) Tumor-related protein, par... 374 e-105
TC15095 similar to UP|AAQ96377 (AAQ96377) Miraculin-like protein... 168 6e-43
BU494288 119 3e-28
TC14855 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precurs... 100 2e-22
TC15508 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precurs... 95 8e-21
TC15509 weakly similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor ... 93 4e-20
TC15888 weakly similar to UP|Q8LK19 (Q8LK19) Proteinase inhibito... 75 1e-14
TC8377 similar to UP|Q9M3Z7 (Q9M3Z7) Alpha-fucosidase , partial... 68 1e-12
TC11107 63 4e-11
TC13211 weakly similar to UP|Q8LK19 (Q8LK19) Proteinase inhibito... 57 3e-09
TC11805 weakly similar to SP|Q42208|RL7_ARATH 60S ribosomal prot... 30 0.24
TC17300 similar to UP|Q9LDS6 (Q9LDS6) Serine/threonine kinase-li... 27 2.7
TC14194 weakly similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specific... 26 5.9
AV779413 26 5.9
BP038323 26 5.9
>TC18413 similar to UP|P93378 (P93378) Tumor-related protein, partial (41%)
Length = 740
Score = 374 bits (961), Expect = e-105
Identities = 173/213 (81%), Positives = 191/213 (89%)
Frame = -3
Query: 1 MKSICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPC 60
MKSI + A+LFAL TQP LG+ DASPEQVVDT+GK VRAG+ YYIRPVPTTPCDGRGPC
Sbjct: 714 MKSIYLSFAILFALCTQPFLGKGDASPEQVVDTQGKNVRAGMGYYIRPVPTTPCDGRGPC 535
Query: 61 VVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSC 120
VVGSGFVLIARSPN+TCPLNVVVVEGFRGQ VTFTPVNPKKGVIRVSTD+NI +L T+C
Sbjct: 534 VVGSGFVLIARSPNKTCPLNVVVVEGFRGQAVTFTPVNPKKGVIRVSTDVNINVTLETAC 355
Query: 121 EESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCK 180
EEST+W LDDFD+S G WFVTTGGV+GNPGKDT+ NWFKIEKYE+DYK VFCPTVC+ CK
Sbjct: 354 EESTVWKLDDFDASAGHWFVTTGGVVGNPGKDTISNWFKIEKYEEDYKLVFCPTVCDTCK 175
Query: 181 VMCRNVGIFRDSNGNQRVALTDVPYKVRFQPSA 213
+CRNVG+F DSNGN+RVALTD YKVRFQPSA
Sbjct: 174 PLCRNVGVFMDSNGNRRVALTDESYKVRFQPSA 76
>TC15095 similar to UP|AAQ96377 (AAQ96377) Miraculin-like protein
(Fragment), partial (42%)
Length = 979
Score = 168 bits (426), Expect = 6e-43
Identities = 97/207 (46%), Positives = 133/207 (63%), Gaps = 6/207 (2%)
Frame = +3
Query: 10 VLFALSTQPLLGEADAS-PEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVL 68
+ FA +T+ L+G A A+ PE V+D G+K+R GV YYI PV G G + SG +
Sbjct: 216 ICFAFTTEFLIGIASAAAPEPVLDISGQKLRTGVKYYILPVLRGK--GGGLTLTSSGNI- 386
Query: 69 IARSPNETCPLNVVV--VEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSL-NTSCEESTI 125
N TCPL+VV +E +GQ VTFTP N K GVI STDLNIK+SL NT+C ++ +
Sbjct: 387 ----NNNTCPLHVVQEKLEVLKGQSVTFTPYNAKGGVILTSTDLNIKSSLTNTTCAKAPV 554
Query: 126 WTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKVMCRN 185
W L +G WF+ TGGV GNPG T+ NWFKIEK + DY F FCP+VC C+ +CR
Sbjct: 555 WKL--LKELSGVWFLATGGVEGNPGMATISNWFKIEKADKDYVFSFCPSVCK-CQTLCRE 725
Query: 186 VGIFRDSNGNQRVALTD--VPYKVRFQ 210
+GI+ D G++ ++L+D P+++ F+
Sbjct: 726 LGIY-DYGGDKHLSLSDQVPPFRIMFK 803
>BU494288
Length = 438
Score = 119 bits (299), Expect = 3e-28
Identities = 63/121 (52%), Positives = 77/121 (63%), Gaps = 3/121 (2%)
Frame = +3
Query: 74 NETCPLNVVV--VEGFRGQGVTFTPVNPKKGVIRVSTDLNIKT-SLNTSCEESTIWTLDD 130
N++CP NVV +E + G VTFTP N K GVI STDLNIK+ + + C S +W L
Sbjct: 30 NKSCPPNVVQAKLEVWNGTPVTFTPYNAKDGVILTSTDLNIKSFPIKSPCGPSFVWKLQK 209
Query: 131 FDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKVMCRNVGIFR 190
TG WF+ TGGV GNPG DT+ NWFKIEK DY FCP+VC C +CR +GI+
Sbjct: 210 --ELTGVWFLVTGGVEGNPGADTIVNWFKIEKAGKDYVLSFCPSVCK-CNTLCRELGIYI 380
Query: 191 D 191
D
Sbjct: 381 D 383
>TC14855 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precursor, partial
(90%)
Length = 1001
Score = 100 bits (249), Expect = 2e-22
Identities = 71/211 (33%), Positives = 107/211 (50%), Gaps = 8/211 (3%)
Frame = +1
Query: 11 LFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIA 70
L L+TQPLL A+ PE VVD +G + GV YY P+ +G + +
Sbjct: 85 LLILNTQPLLA-AEPEPEPVVDKQGNPLEPGVGYYAWPL-----------WADNGGLTLG 228
Query: 71 RSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTS-LNTSCEESTIWTLD 129
R+ N+TCPL+++ G + F + G I TDL ++ L + C+E +W L
Sbjct: 229 RTRNKTCPLDIIRDPSKIGTPIEFHVADSDLGYIPTLTDLTVEMPILGSRCQEPKVWRL- 405
Query: 130 DFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDD-----YKFVFCPTVCNFCKVMCR 184
+G WF++TGGV GN + + FKIE+ + Y F FCP+V V+C
Sbjct: 406 -LKVGSGFWFLSTGGVAGN-----LVSKFKIERLAGEHAYEIYSFKFCPSVPG---VLCA 558
Query: 185 NVGIFRDSNGNQRVALTD--VPYKVRFQPSA 213
VG + D++G Q +A+ D Y VRFQ ++
Sbjct: 559 PVGTYEDADGTQVMAIGDDIETYYVRFQKAS 651
>TC15508 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precursor, partial
(66%)
Length = 877
Score = 95.1 bits (235), Expect = 8e-21
Identities = 74/215 (34%), Positives = 105/215 (48%), Gaps = 15/215 (6%)
Frame = +3
Query: 11 LFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIA 70
L L+TQPLL AD VVD +G + GV YY+RPV G + +
Sbjct: 69 LLVLNTQPLL--ADEPEPAVVDMQGNPLLPGVQYYVRPVSAE-----------KGGLALG 209
Query: 71 RSPNETCPLNVVV-VEGFRGQGVTF-TPVNPKKGVIRVSTDLNIKTS-LNTSCEESTIWT 127
+ N+TCPL++++ G F T + G I +STDL ++ L + C+E +W
Sbjct: 210 HTRNKTCPLDIILDPSSIIGSPTVFHTSSSSDVGYIPISTDLTVEMPILGSPCKEPKVWR 389
Query: 128 LDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDD---------YKFVFCPTVCNF 178
L S WFV+T GV GN + + FKI ++E+ Y F FCP+V
Sbjct: 390 LAKVGSDF--WFVSTQGVPGN-----LVSKFKIVRFEERDNHANEGEIYSFSFCPSVYG- 545
Query: 179 CKVMCRNVGIFRDSNGNQRVAL---TDVPYKVRFQ 210
V+C VGI+ D +G Q +A+ PY VRFQ
Sbjct: 546 --VICAPVGIYVDDDGTQVMAVGAGVGKPYHVRFQ 644
>TC15509 weakly similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precursor,
partial (77%)
Length = 797
Score = 92.8 bits (229), Expect = 4e-20
Identities = 68/214 (31%), Positives = 103/214 (47%), Gaps = 14/214 (6%)
Frame = +1
Query: 11 LFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIA 70
L L+TQPLL AD VD +G + GV YY+RPV +G + +
Sbjct: 46 LLVLNTQPLL--ADEPEPAAVDMQGNPILPGVKYYVRPVSAE-----------NGGLALG 186
Query: 71 RSPNETCPLNVVVVEGFRGQGVTF-TPVNPKKGVIRVSTDLNIKTS-LNTSCEESTIWTL 128
+ N+TCPL++++ G F T + G I +STDL ++ L + C+E +W +
Sbjct: 187 HTRNKTCPLDIILDPFSLGTPTVFQTSSSSDLGYIPISTDLTVEMPVLKSPCKEPKVWRI 366
Query: 129 DDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDD---------YKFVFCPTVCNFC 179
WFV+T GV GN + FKI ++E+ Y F +CP++ +
Sbjct: 367 --AKEGADFWFVSTQGVPGNDA-----SRFKIVRFEEGDNHANEGEIYSFSYCPSIWGY- 522
Query: 180 KVMCRNVGIFRDSNGNQRVAL---TDVPYKVRFQ 210
+C VGI+ D +G Q +A+ PY VRFQ
Sbjct: 523 --ICAPVGIYVDDDGTQVMAVGAGVGKPYHVRFQ 618
>TC15888 weakly similar to UP|Q8LK19 (Q8LK19) Proteinase inhibitor 20,
partial (27%)
Length = 611
Score = 74.7 bits (182), Expect = 1e-14
Identities = 68/196 (34%), Positives = 91/196 (45%), Gaps = 6/196 (3%)
Frame = +3
Query: 7 LLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGF 66
L +LFA +T + P+ V DT G +R+ YYIRP RGP G G
Sbjct: 69 LSVLLFAFTTYIIPSAFSQVPDLVKDTNGIPLRSDGRYYIRPAL------RGP---GGGG 221
Query: 67 VLIARSPNETCPLNVV--VVEGFRGQGVTFTPVNPKKGVIRVSTDLNIK-TSLNTSCEES 123
V + N+TCP+ V+ E G GV F P G+I L I + C +S
Sbjct: 222 VSPGETGNQTCPVTVLQHKSEVENGNGVKFEHY-PNIGIIYEGVPLTISFGDIYGFCADS 398
Query: 124 TIWTL--DDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYED-DYKFVFCPTVCNFCK 180
T W + DDF G+W V GG +PGK+ V F I+KY+D YK VF P V
Sbjct: 399 TKWVVVSDDF---PGKW-VGIGGAEDHPGKEIVSGLFDIQKYDDVAYKLVFTP-VTGGSP 563
Query: 181 VMCRNVGIFRDSNGNQ 196
+ ++G D NG +
Sbjct: 564 GLSVDIGRRDDKNGRR 611
>TC8377 similar to UP|Q9M3Z7 (Q9M3Z7) Alpha-fucosidase , partial (83%)
Length = 1014
Score = 67.8 bits (164), Expect = 1e-12
Identities = 64/213 (30%), Positives = 94/213 (44%), Gaps = 11/213 (5%)
Frame = +1
Query: 1 MKSICILLAVLFA--LSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRG 58
MK I + L LF +ST+ + EQV+D G + G YYIRP RG
Sbjct: 31 MKPIPLSLCFLFFAFISTKLPTAFSSNDVEQVLDVNGDSIFPGGRYYIRPAI------RG 192
Query: 59 PCVVGSGFVLIARSPNETCPLNVV--VVEGFRGQGVTF-TPVNPKKGVIRVSTDLNIKTS 115
P G V + ++ + CP+ + E G V F G+I T+L+I+ +
Sbjct: 193 P---PGGGVRLGKTGDSDCPVTALQEYSEVKNGIPVRFRIAAEVSTGIIFTGTELDIEFT 363
Query: 116 LNTSCEESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYED--DYKFVFC- 172
+ C ES W + D+ G+ V GG +PGK T F I+K YK VFC
Sbjct: 364 VKPHCVESPKWIV-FVDNEIGKACVGIGGAKDHPGKQTFSGTFNIQKNSAGFGYKLVFCI 540
Query: 173 ---PTVCNFCKVMCRNVGIFRDSNGNQRVALTD 202
PT C ++G + + G +R+ LT+
Sbjct: 541 KGSPT--------CLDIGRYDNGEGGRRLNLTE 615
>TC11107
Length = 841
Score = 62.8 bits (151), Expect = 4e-11
Identities = 41/132 (31%), Positives = 61/132 (46%), Gaps = 1/132 (0%)
Frame = +3
Query: 80 NVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTIWTLDDFDSSTGQWF 139
N + EGF V FTP +++ D + S T+C +ST W++ ++ +G+
Sbjct: 6 NTDLPEGFP---VKFTPFAKGDKYVKLGKDFTVVFSAATTCVQSTAWSVGSTEAKSGRRL 176
Query: 140 VTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPT-VCNFCKVMCRNVGIFRDSNGNQRV 198
V TGG +G + +E Y +CPT VCN C+ C GI R+ NG +
Sbjct: 177 VVTGGDVGERAYGSYFRIVAVEGRAGIYNIQWCPTDVCNICRFRCGTAGILRE-NGKILL 353
Query: 199 ALTDVPYKVRFQ 210
AL V FQ
Sbjct: 354 ALDGGMLPVVFQ 389
>TC13211 weakly similar to UP|Q8LK19 (Q8LK19) Proteinase inhibitor 20,
partial (44%)
Length = 558
Score = 56.6 bits (135), Expect = 3e-09
Identities = 58/183 (31%), Positives = 79/183 (42%), Gaps = 11/183 (6%)
Frame = +1
Query: 1 MKSICILLAVLFALSTQPLLGEADAS--PEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRG 58
MK +L F + P L S PEQV D G + G Y I P RG
Sbjct: 19 MKPTILLSLCFFLFALTPYLFPLAFSQDPEQVKDAYGNPLVPGRRYSIVPAL------RG 180
Query: 59 PCVVGSGFVLIARSPNETCPLNVV--VVEGFRGQGVTFTP--VNPKKGVIRVSTDLNIKT 114
SG V ++ N+TCPL VV E G+ V F+ + P G I T L+I
Sbjct: 181 S---RSGGVSPDKTGNQTCPLTVVQEYFECRDGKPVIFSVSGITPGTGTILTGTHLDIAF 351
Query: 115 SLNTSCEESTIWT----LDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYE-DDYKF 169
C +++ W L D+ + V+ GGV +PG+ +D F I+K + YK
Sbjct: 352 VEKPECADTSKWVVVGNLTDYPGAP----VSIGGVEDHPGQHILDGTFNIQKLDCGGYKL 519
Query: 170 VFC 172
VFC
Sbjct: 520 VFC 528
>TC11805 weakly similar to SP|Q42208|RL7_ARATH 60S ribosomal protein L7.
{Arabidopsis thaliana;}, partial (43%)
Length = 829
Score = 30.4 bits (67), Expect = 0.24
Identities = 17/57 (29%), Positives = 28/57 (48%), Gaps = 6/57 (10%)
Frame = -2
Query: 94 FTPVNPKKGVIRVSTDLNIKTSL------NTSCEESTIWTLDDFDSSTGQWFVTTGG 144
F P NP G++R +N+ TSL + SC S++ T+ + SS ++ G
Sbjct: 729 FDPFNPSDGLLRSKGHINLTTSLKWGPVFSISCTMSSMHTIPNLPSSCSMILLSVKG 559
>TC17300 similar to UP|Q9LDS6 (Q9LDS6) Serine/threonine kinase-like protein
, partial (6%)
Length = 626
Score = 26.9 bits (58), Expect = 2.7
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 1/47 (2%)
Frame = -2
Query: 103 VIRVSTDLNIK-TSLNTSCEESTIWTLDDFDSSTGQWFVTTGGVLGN 148
++ +ST +I+ SL SC + + T + F SSTG + TG + N
Sbjct: 274 LVSISTTSDIRGRSLAVSCTQRSPTTRNRFASSTGIFIFNTGSTMLN 134
>TC14194 weakly similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specific fungal
endoglucanase inhibitor protein precursor, partial (11%)
Length = 892
Score = 25.8 bits (55), Expect = 5.9
Identities = 11/34 (32%), Positives = 15/34 (43%)
Frame = +3
Query: 48 PVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNV 81
P T C PC+ +GF N TC L++
Sbjct: 282 PCDTQKCPQNSPCIGCNGFPTKPGCTNNTCGLSI 383
>AV779413
Length = 511
Score = 25.8 bits (55), Expect = 5.9
Identities = 9/22 (40%), Positives = 13/22 (58%)
Frame = +1
Query: 48 PVPTTPCDGRGPCVVGSGFVLI 69
PVP C G P V+G+ F ++
Sbjct: 163 PVPKVSCVGNDPSVIGTAFYIM 228
>BP038323
Length = 453
Score = 25.8 bits (55), Expect = 5.9
Identities = 16/43 (37%), Positives = 21/43 (48%)
Frame = -3
Query: 42 VDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVV 84
V YYI+PV +G G VLI +P+E PL V +
Sbjct: 274 VSYYIKPV------------LGEGQVLILSAPHEEGPLGRVAL 182
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,145,241
Number of Sequences: 28460
Number of extensions: 58460
Number of successful extensions: 283
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 273
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 273
length of query: 213
length of database: 4,897,600
effective HSP length: 86
effective length of query: 127
effective length of database: 2,450,040
effective search space: 311155080
effective search space used: 311155080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 53 (25.0 bits)
Medicago: description of AC140022.13


