

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.3 + phase: 0
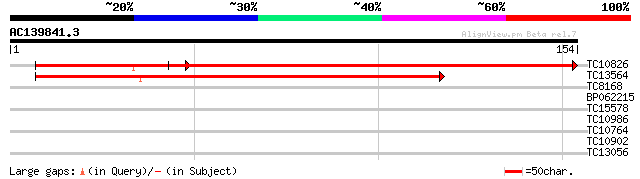
(154 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC10826 similar to UP|RL2B_FRIAG (O22644) 60S ribosomal protein ... 206 3e-66
TC13564 similar to UP|Q9LWB3 (Q9LWB3) Ribosomal protein L23, par... 200 1e-52
TC8168 similar to UP|O04720 (O04720) Cysteine proteinase inhibit... 34 0.013
BP062215 27 1.5
TC15578 26 2.6
TC10986 similar to GB|AAL56097.1|18028661|AF334030 ORF91 {Helico... 26 3.4
TC10764 25 4.5
TC10902 similar to UP|O04794 (O04794) Acyl-ACP thioesterase, par... 25 5.9
TC13056 similar to UP|IF38_MEDTR (Q9XHM1) Eukaryotic translation... 25 7.7
>TC10826 similar to UP|RL2B_FRIAG (O22644) 60S ribosomal protein L23A,
partial (94%)
Length = 754
Score = 206 bits (524), Expect(2) = 3e-66
Identities = 103/111 (92%), Positives = 106/111 (94%)
Frame = +1
Query: 44 FHRPKTQTKDRNPKYPRISATPRNKLDHYQILKFPLTTESAMKKIEDNNTLVFIVDLRAD 103
F P+ TK+RNPKYPRISATPRNKLDHYQILKFPLTTESAMKKIEDNNTLVFIVD+RAD
Sbjct: 211 FTGPRHCTKERNPKYPRISATPRNKLDHYQILKFPLTTESAMKKIEDNNTLVFIVDIRAD 390
Query: 104 KKKIKDAVKKMYDIQAKKVNTLIRPDGTKKAYVRLTPDYDALDVANKIGII 154
KKKIK AVKKMYDIQAKKVNTLIRPDGTKKAYVRLTPDYDALDVANKIGII
Sbjct: 391 KKKIKAAVKKMYDIQAKKVNTLIRPDGTKKAYVRLTPDYDALDVANKIGII 543
Score = 60.8 bits (146), Expect(2) = 3e-66
Identities = 32/44 (72%), Positives = 34/44 (76%), Gaps = 2/44 (4%)
Frame = +3
Query: 8 DVSKKTDPKAQALKAAKAVKSGTAI--KKAKKIRTTVTFHRPKT 49
D +KK D KAQAL AKAVKSG + KKA KIRTTVTFHRPKT
Sbjct: 96 DTTKKADAKAQALNTAKAVKSGGKVIKKKANKIRTTVTFHRPKT 227
>TC13564 similar to UP|Q9LWB3 (Q9LWB3) Ribosomal protein L23, partial (74%)
Length = 396
Score = 200 bits (508), Expect = 1e-52
Identities = 104/112 (92%), Positives = 104/112 (92%), Gaps = 1/112 (0%)
Frame = +3
Query: 8 DVSKKTDPKAQALKAAKAVKSGTAIKK-AKKIRTTVTFHRPKTQTKDRNPKYPRISATPR 66
D SKK DPKAQALKAAKAVKSGT IKK KKIRTTVTFHRPKT TK RNPKYPRISATPR
Sbjct: 60 DSSKKNDPKAQALKAAKAVKSGTTIKKKTKKIRTTVTFHRPKTLTKTRNPKYPRISATPR 239
Query: 67 NKLDHYQILKFPLTTESAMKKIEDNNTLVFIVDLRADKKKIKDAVKKMYDIQ 118
NKLDHYQILKFPLTTESAMKKIEDNNTLVFIVDLRADKKKIK AVKKMYDIQ
Sbjct: 240 NKLDHYQILKFPLTTESAMKKIEDNNTLVFIVDLRADKKKIKAAVKKMYDIQ 395
>TC8168 similar to UP|O04720 (O04720) Cysteine proteinase inhibitor,
partial (55%)
Length = 808
Score = 33.9 bits (76), Expect = 0.013
Identities = 22/72 (30%), Positives = 35/72 (48%), Gaps = 5/72 (6%)
Frame = +3
Query: 2 HCFCAGDVSKKTDP-----KAQALKAAKAVKSGTAIKKAKKIRTTVTFHRPKTQTKDRNP 56
HC A + SKK +L + + S T I +IR+ V H+ + R+P
Sbjct: 165 HCHSASNQSKKPHEIFFFHSLLSLPLLRPLHSSTIICCLFRIRSRVNGHQAR-----RHP 329
Query: 57 KYPRISATPRNK 68
++PR+S PRN+
Sbjct: 330 RFPRVSELPRNR 365
>BP062215
Length = 314
Score = 26.9 bits (58), Expect = 1.5
Identities = 13/41 (31%), Positives = 20/41 (48%), Gaps = 4/41 (9%)
Frame = +1
Query: 36 KKIRTTVTFHRPKTQTK----DRNPKYPRISATPRNKLDHY 72
+K +T H+P TK D PK+P +P++ HY
Sbjct: 178 QKTAMRLTHHQPLLSTKK*QLDNIPKHPHYLKSPKHSSPHY 300
>TC15578
Length = 667
Score = 26.2 bits (56), Expect = 2.6
Identities = 10/19 (52%), Positives = 15/19 (78%)
Frame = -2
Query: 54 RNPKYPRISATPRNKLDHY 72
R+PK+P+ SAT + +L HY
Sbjct: 72 RHPKHPKSSATRKVQLHHY 16
>TC10986 similar to GB|AAL56097.1|18028661|AF334030 ORF91 {Helicoverpa zea
single nucleocapsid nucleopolyhedrovirus;}, partial
(15%)
Length = 655
Score = 25.8 bits (55), Expect = 3.4
Identities = 10/27 (37%), Positives = 16/27 (59%)
Frame = -3
Query: 43 TFHRPKTQTKDRNPKYPRISATPRNKL 69
T+H+P+ Q ++ NP + R A R L
Sbjct: 245 TYHKPRKQKREENPIFGREKAKWRGNL 165
>TC10764
Length = 555
Score = 25.4 bits (54), Expect = 4.5
Identities = 10/29 (34%), Positives = 18/29 (61%)
Frame = -2
Query: 5 CAGDVSKKTDPKAQALKAAKAVKSGTAIK 33
C +SK+ DPK + + +A ++ +AIK
Sbjct: 275 CK*PISKQNDPKHEVISEERAQQNASAIK 189
>TC10902 similar to UP|O04794 (O04794) Acyl-ACP thioesterase, partial (11%)
Length = 810
Score = 25.0 bits (53), Expect = 5.9
Identities = 8/20 (40%), Positives = 11/20 (55%)
Frame = -2
Query: 53 DRNPKYPRISATPRNKLDHY 72
D+ P YP + P N + HY
Sbjct: 677 DKAPTYPNLPHKPNNNMHHY 618
>TC13056 similar to UP|IF38_MEDTR (Q9XHM1) Eukaryotic translation initiation
factor 3 subunit 8 (eIF3 p110) (eIF3c), partial (12%)
Length = 607
Score = 24.6 bits (52), Expect = 7.7
Identities = 13/32 (40%), Positives = 17/32 (52%)
Frame = +2
Query: 94 LVFIVDLRADKKKIKDAVKKMYDIQAKKVNTL 125
LV + D A+ KDA KKM AK +N +
Sbjct: 476 LVMLEDFLAEALANKDAKKKMSSSNAKALNAM 571
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.132 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,975,354
Number of Sequences: 28460
Number of extensions: 17659
Number of successful extensions: 101
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 99
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 99
length of query: 154
length of database: 4,897,600
effective HSP length: 83
effective length of query: 71
effective length of database: 2,535,420
effective search space: 180014820
effective search space used: 180014820
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 51 (24.3 bits)
Medicago: description of AC139841.3


