

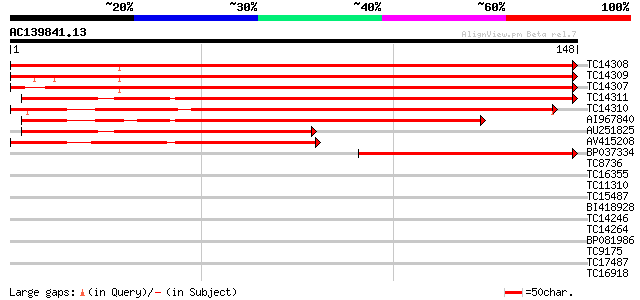
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.13 - phase: 0
(148 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14308 homologue to UP|H2B_GOSHI (O22582) Histone H2B, partial ... 275 2e-75
TC14309 homologue to UP|H2B_GOSHI (O22582) Histone H2B, partial ... 267 6e-73
TC14307 homologue to UP|H2B_GOSHI (O22582) Histone H2B, partial ... 260 6e-71
TC14311 homologue to UP|Q9FFC0 (Q9FFC0) Histone H2B like protein... 232 2e-62
TC14310 homologue to UP|Q9M3H6 (Q9M3H6) Histone H2B, partial (92%) 217 8e-58
AI967840 171 4e-44
AU251825 114 9e-27
AV415208 112 2e-26
BP037334 88 5e-19
TC8736 similar to PIR|T06396|T06396 isoprenylated protein - soyb... 37 0.002
TC16355 similar to UP|Q9M4U6 (Q9M4U6) Histone H1 variant, partia... 36 0.002
TC11310 similar to UP|BAD11331 (BAD11331) BRI1-KD interacting pr... 34 0.009
TC15487 similar to UP|O23144 (O23144) Proton pump interactor (AT... 34 0.012
BI418928 33 0.015
TC14246 similar to UP|Q84LL6 (Q84LL6) Salt tolerance protein 5, ... 31 0.077
TC14264 similar to UP|Q84NF9 (Q84NF9) Histone H1 (Fragment), par... 31 0.10
BP081986 31 0.10
TC9175 weakly similar to UP|INAD_DROME (Q24008) Inactivation-no-... 31 0.10
TC17487 UP|CAE45597 (CAE45597) SAR DNA-binding protein-like prot... 30 0.13
TC16918 weakly similar to UP|Q9FJH5 (Q9FJH5) Gb|AAC98457.1 (At5g... 30 0.13
>TC14308 homologue to UP|H2B_GOSHI (O22582) Histone H2B, partial (96%)
Length = 764
Score = 275 bits (703), Expect = 2e-75
Identities = 143/149 (95%), Positives = 146/149 (97%), Gaps = 1/149 (0%)
Frame = +3
Query: 1 MAPKGEKKPAEKKPAEEKKSTVAEKAP-AEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVE 59
MAPK +KKPAEKKPAEEKKSTVAEKAP AEKKPKAGKKLPKEG +AAGDKKKKRSKK+VE
Sbjct: 75 MAPKADKKPAEKKPAEEKKSTVAEKAPPAEKKPKAGKKLPKEGAAAAGDKKKKRSKKSVE 254
Query: 60 TYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQ 119
TYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQ
Sbjct: 255 TYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQ 434
Query: 120 TAVRLVLPGELAKHAVSEGTKAVTKFTSS 148
TAVRLVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 435 TAVRLVLPGELAKHAVSEGTKAVTKFTSS 521
>TC14309 homologue to UP|H2B_GOSHI (O22582) Histone H2B, partial (93%)
Length = 682
Score = 267 bits (682), Expect = 6e-73
Identities = 144/152 (94%), Positives = 145/152 (94%), Gaps = 4/152 (2%)
Frame = +1
Query: 1 MAPKG--EKKPA-EKKPAEEKKSTVAEKAP-AEKKPKAGKKLPKEGGSAAGDKKKKRSKK 56
MAPK EKKPA EKKPAEEKKST AEKAP AEKKPKAGKKLPKEG SAAGDKKKKRSKK
Sbjct: 58 MAPKAAAEKKPAAEKKPAEEKKSTAAEKAPPAEKKPKAGKKLPKEGASAAGDKKKKRSKK 237
Query: 57 NVETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSR 116
NVETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSR
Sbjct: 238 NVETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSR 417
Query: 117 EIQTAVRLVLPGELAKHAVSEGTKAVTKFTSS 148
EIQTAVRLVLPGELAKHAVSEGTK+VTKFTSS
Sbjct: 418 EIQTAVRLVLPGELAKHAVSEGTKSVTKFTSS 513
>TC14307 homologue to UP|H2B_GOSHI (O22582) Histone H2B, partial (90%)
Length = 701
Score = 260 bits (665), Expect = 6e-71
Identities = 139/149 (93%), Positives = 142/149 (95%), Gaps = 1/149 (0%)
Frame = +2
Query: 1 MAPKGEKKPAEKKPAEEKKSTVAEKAP-AEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVE 59
MAPK A+KKPAEEKKSTVAEKAP AEKKPKAGKKLPKEG +AAGDKKKKRSKK+VE
Sbjct: 80 MAPK-----ADKKPAEEKKSTVAEKAPPAEKKPKAGKKLPKEGAAAAGDKKKKRSKKSVE 244
Query: 60 TYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQ 119
TYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQ
Sbjct: 245 TYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQ 424
Query: 120 TAVRLVLPGELAKHAVSEGTKAVTKFTSS 148
TAVRLVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 425 TAVRLVLPGELAKHAVSEGTKAVTKFTSS 511
>TC14311 homologue to UP|Q9FFC0 (Q9FFC0) Histone H2B like protein, partial
(97%)
Length = 626
Score = 232 bits (591), Expect = 2e-62
Identities = 122/145 (84%), Positives = 133/145 (91%)
Frame = +1
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVETYKI 63
+ +KKPAEKKPA ++ A AEKKP+A KK+PKEG +AAGDKKKKRSKKNVETYKI
Sbjct: 49 RADKKPAEKKPAAAEEKAAA----AEKKPRAEKKIPKEG-AAAGDKKKKRSKKNVETYKI 213
Query: 64 YIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQTAVR 123
YIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYN+KPTITSREIQTAVR
Sbjct: 214 YIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNQKPTITSREIQTAVR 393
Query: 124 LVLPGELAKHAVSEGTKAVTKFTSS 148
LVLPGELA+HAVSEGT+AVT+FTSS
Sbjct: 394 LVLPGELAQHAVSEGTQAVTQFTSS 468
>TC14310 homologue to UP|Q9M3H6 (Q9M3H6) Histone H2B, partial (92%)
Length = 502
Score = 217 bits (552), Expect = 8e-58
Identities = 122/145 (84%), Positives = 127/145 (87%), Gaps = 2/145 (1%)
Frame = +3
Query: 1 MAP-KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVE 59
MAP K EKKPAEKKPA AEKAPAEKKPKA KK+ KEGGS +KKKK+SKK+VE
Sbjct: 57 MAPAKAEKKPAEKKPA-------AEKAPAEKKPKAEKKIAKEGGS---EKKKKKSKKSVE 206
Query: 60 TYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQ 119
TYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQ
Sbjct: 207 TYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQ 386
Query: 120 TAVRLVLPGELAKHAVSEGTK-AVT 143
TAVRL LPGELAKHAVSE + AVT
Sbjct: 387 TAVRLGLPGELAKHAVSEXNQXAVT 461
>AI967840
Length = 393
Score = 171 bits (434), Expect = 4e-44
Identities = 96/121 (79%), Positives = 102/121 (83%)
Frame = +1
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVETYKI 63
KGEKKPAEKKPA AEKAP K A KK+P + +A+ DKKKK+ K+VETYKI
Sbjct: 58 KGEKKPAEKKPA-------AEKAPPAK---AEKKVPTKD-AASVDKKKKKKSKSVETYKI 204
Query: 64 YIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQTAVR 123
YIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQTAVR
Sbjct: 205 YIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQTAVR 384
Query: 124 L 124
L
Sbjct: 385 L 387
>AU251825
Length = 237
Score = 114 bits (284), Expect = 9e-27
Identities = 58/77 (75%), Positives = 64/77 (82%)
Frame = +3
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVETYKI 63
KG+KKPAEKKPA ++ A AEKKP+A KK+P + G AAGDKKKKRSKKNVETYKI
Sbjct: 18 KGDKKPAEKKPAAAEEKAAA----AEKKPRAEKKIPAKEGGAAGDKKKKRSKKNVETYKI 185
Query: 64 YIFKVLKQVHPDIGISS 80
YIFKVLKQVHPDIGISS
Sbjct: 186YIFKVLKQVHPDIGISS 236
>AV415208
Length = 242
Score = 112 bits (281), Expect = 2e-26
Identities = 60/81 (74%), Positives = 66/81 (81%)
Frame = +3
Query: 1 MAPKGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVET 60
MAPK EKKPAEKKPA AEKAPAEKKPKA KK+ K+ + +KKKKR+KK+VET
Sbjct: 24 MAPKAEKKPAEKKPA------AAEKAPAEKKPKAEKKISKD--ATGSEKKKKRTKKSVET 179
Query: 61 YKIYIFKVLKQVHPDIGISSK 81
YKIYIFKVLKQVHPDIGISSK
Sbjct: 180YKIYIFKVLKQVHPDIGISSK 242
>BP037334
Length = 318
Score = 88.2 bits (217), Expect = 5e-19
Identities = 46/57 (80%), Positives = 51/57 (88%)
Frame = -2
Query: 92 DIFEKLAQESSRLARYNKKPTITSREIQTAVRLVLPGELAKHAVSEGTKAVTKFTSS 148
DIFEKLAQESSRLARY++KPTITSREI VRLVLPGELA+ VSEG +AVT+FTSS
Sbjct: 317 DIFEKLAQESSRLARYHQKPTITSREIPPPVRLVLPGELAQPPVSEGPQAVTQFTSS 147
>TC8736 similar to PIR|T06396|T06396 isoprenylated protein - soybean
(fragment) {Glycine max;}, partial (84%)
Length = 1268
Score = 36.6 bits (83), Expect = 0.002
Identities = 19/48 (39%), Positives = 25/48 (51%)
Frame = +2
Query: 5 GEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKK 52
GEK A +P ++K S + A EKK K+ PK G GD+K K
Sbjct: 542 GEKPKAAAEPEKKKDSEKPKAAEPEKKKDGDKEKPKSDGPKKGDEKPK 685
>TC16355 similar to UP|Q9M4U6 (Q9M4U6) Histone H1 variant, partial (50%)
Length = 584
Score = 36.2 bits (82), Expect = 0.002
Identities = 19/30 (63%), Positives = 20/30 (66%)
Frame = +1
Query: 7 KKPAEKKPAEEKKSTVAEKAPAEKKPKAGK 36
K P E KPA+EKK KAP EKKPK GK
Sbjct: 121 KAPKETKPAKEKK----PKAPKEKKPKQGK 198
Score = 35.0 bits (79), Expect = 0.005
Identities = 20/39 (51%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Frame = +1
Query: 9 PAEKKPAEEKKSTVAEKAPAEKKPKAGK-KLPKEGGSAA 46
PA +KP EE K+ K EKKPKA K K PK+G A+
Sbjct: 91 PAVEKPVEEVKAPKETKPAKEKKPKAPKEKKPKQGKIAS 207
Score = 24.6 bits (52), Expect = 7.2
Identities = 16/47 (34%), Positives = 20/47 (42%)
Frame = +1
Query: 17 EKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVETYKI 63
E + V PA +KP K PKE A K K +K + KI
Sbjct: 61 EMTAAVESVTPAVEKPVEEVKAPKETKPAKEKKPKAPKEKKPKQGKI 201
>TC11310 similar to UP|BAD11331 (BAD11331) BRI1-KD interacting protein 102
(Fragment), partial (13%)
Length = 667
Score = 34.3 bits (77), Expect = 0.009
Identities = 19/53 (35%), Positives = 29/53 (53%)
Frame = +1
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKK 56
K EKK +KK E + +++ AEK+ K K+ E GS D+ +K+ KK
Sbjct: 82 KKEKKKKKKKDQENGEVASSDEEKAEKEKKKKHKVKVEDGSPDLDQSEKKKKK 240
>TC15487 similar to UP|O23144 (O23144) Proton pump interactor
(AT4g27500/F27G19_100), partial (16%)
Length = 1153
Score = 33.9 bits (76), Expect = 0.012
Identities = 22/53 (41%), Positives = 29/53 (54%)
Frame = +2
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKK 56
K E++ A+ K A E+K +AEKA A KA K KE D++KK KK
Sbjct: 452 KREEEIAKAKQAWERKKKLAEKAAA----KAALKAQKEAEKKLKDREKKAKKK 598
>BI418928
Length = 496
Score = 33.5 bits (75), Expect = 0.015
Identities = 16/39 (41%), Positives = 21/39 (53%)
Frame = +3
Query: 3 PKGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKE 41
P EKKP EK ++KK + P EKK + K PK+
Sbjct: 282 PPPEKKPEEKPKTDDKKPEEEKTKPDEKKAEEKTKTPKQ 398
Score = 32.0 bits (71), Expect = 0.045
Identities = 21/59 (35%), Positives = 30/59 (50%)
Frame = +3
Query: 5 GEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVETYKI 63
G+K P EKKP E+ K+ +K P E+K K +K +E K K K++ KI
Sbjct: 273 GDKPPPEKKPEEKPKTD--DKKPEEEKTKPDEKKAEE--------KTKTPKQSTVVMKI 419
>TC14246 similar to UP|Q84LL6 (Q84LL6) Salt tolerance protein 5, partial
(57%)
Length = 1280
Score = 31.2 bits (69), Expect = 0.077
Identities = 17/40 (42%), Positives = 22/40 (54%)
Frame = +1
Query: 6 EKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSA 45
+K+ E K +EEK AEKKPKA + KEG +A
Sbjct: 352 KKREEELKASEEKAKAEKRLKEAEKKPKAEENDKKEGTAA 471
Score = 28.5 bits (62), Expect = 0.50
Identities = 17/40 (42%), Positives = 22/40 (54%)
Frame = +1
Query: 12 KKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKK 51
KK EE+ EKA AEK+ K +K PK + DKK+
Sbjct: 349 KKKREEELKASEEKAKAEKRLKEAEKKPK---AEENDKKE 459
>TC14264 similar to UP|Q84NF9 (Q84NF9) Histone H1 (Fragment), partial (76%)
Length = 1277
Score = 30.8 bits (68), Expect = 0.10
Identities = 17/45 (37%), Positives = 24/45 (52%), Gaps = 6/45 (13%)
Frame = +2
Query: 5 GEKKPAEKKPAEEKKSTVAEKAPAEK------KPKAGKKLPKEGG 43
G+K P KPA +K +T +KAPA+ K A + P+ GG
Sbjct: 881 GKKVPPPPKPAPKKAATPVKKAPAKSGKAKTVKSPAKRTAPQRGG 1015
Score = 28.9 bits (63), Expect = 0.38
Identities = 15/33 (45%), Positives = 17/33 (51%)
Frame = +2
Query: 2 APKGEKKPAEKKPAEEKKSTVAEKAPAEKKPKA 34
A K + KPA K A K+T K A KPKA
Sbjct: 563 ATKAKAKPAPKSKAAATKTTAKAKPAAAAKPKA 661
Score = 26.6 bits (57), Expect = 1.9
Identities = 32/145 (22%), Positives = 55/145 (37%), Gaps = 1/145 (0%)
Frame = +2
Query: 1 MAPKGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVET 60
+AP ++P ++PA VAE PA+ P K+ SAA KK S + E
Sbjct: 89 IAPIIVEEPVVEEPAAADLPPVAEDEPAQPDPNPTKEAKPRKRSAA----KKASHPSYEE 256
Query: 61 YKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYN-KKPTITSREIQ 119
LK+ S + + F+ + ++L +L ++ KK + ++
Sbjct: 257 MVKDAIVTLKEK------SGSSQYAIAKFLEEKHKQLPSNFKKLLLFHLKKLVAAGKLVK 418
Query: 120 TAVRLVLPGELAKHAVSEGTKAVTK 144
LP + A + A K
Sbjct: 419 VKNSFKLPSAKSSAAAAASKPAAAK 493
Score = 26.2 bits (56), Expect = 2.5
Identities = 18/52 (34%), Positives = 22/52 (41%)
Frame = +2
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSK 55
K A KPA KK A ++ KPKA K P + A K +SK
Sbjct: 449 KSSAAAAASKPAAAKKKPQPAAAKSKPKPKA-KAKPAAAATKAKAKPAPKSK 601
>BP081986
Length = 433
Score = 30.8 bits (68), Expect = 0.10
Identities = 14/31 (45%), Positives = 19/31 (61%)
Frame = +1
Query: 3 PKGEKKPAEKKPAEEKKSTVAEKAPAEKKPK 33
PK +K P E + + KKST + A AE KP+
Sbjct: 64 PKPKKPPPETQACQIKKSTTQKSASAETKPR 156
>TC9175 weakly similar to UP|INAD_DROME (Q24008)
Inactivation-no-after-potential D protein, partial (5%)
Length = 692
Score = 30.8 bits (68), Expect = 0.10
Identities = 19/57 (33%), Positives = 33/57 (57%), Gaps = 4/57 (7%)
Frame = +2
Query: 1 MAPKGEKKPAEKKPAEEKKSTVAEKA-PAEKKP--KAGKKLPKE-GGSAAGDKKKKR 53
+ P+ E +P +K+ +E++ AE+ P EKK KA ++ KE GG D K+++
Sbjct: 47 IVPQPEPEPEKKEEGKEEEKPAAEEGKPEEKKEEEKAAEEAKKEEGGEEKNDNKEEK 217
Score = 24.3 bits (51), Expect = 9.4
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 3/49 (6%)
Frame = +2
Query: 7 KKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLP---KEGGSAAGDKKKK 52
KK A+ P E + E+ E+KP A + P KE AA + KK+
Sbjct: 32 KKQAKIVPQPEPEPEKKEEGKEEEKPAAEEGKPEEKKEEEKAAEEAKKE 178
>TC17487 UP|CAE45597 (CAE45597) SAR DNA-binding protein-like protein, complete
Length = 1794
Score = 30.4 bits (67), Expect = 0.13
Identities = 18/63 (28%), Positives = 32/63 (50%), Gaps = 11/63 (17%)
Frame = +1
Query: 4 KGEKKPAEKKPAE-----------EKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKK 52
K EKK ++K E +++ V +K +K+ ++ + + + G AG+KKKK
Sbjct: 1534 KKEKKKKKEKNEEKDVPLLADEDGDEEKEVVKKEKKKKRKESTENVELQNGDDAGEKKKK 1713
Query: 53 RSK 55
R K
Sbjct: 1714 RKK 1722
Score = 25.8 bits (55), Expect = 3.2
Identities = 18/54 (33%), Positives = 24/54 (44%)
Frame = +1
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKN 57
K EKK K+ E + + A EKK K K E + KKK + KK+
Sbjct: 1627 KKEKKKKRKESTENVELQNGDDA-GEKKKKRKKHAEPEESAEMPSKKKGKKKKS 1785
>TC16918 weakly similar to UP|Q9FJH5 (Q9FJH5) Gb|AAC98457.1 (At5g60800),
partial (24%)
Length = 498
Score = 30.4 bits (67), Expect = 0.13
Identities = 22/41 (53%), Positives = 23/41 (55%), Gaps = 9/41 (21%)
Frame = +2
Query: 2 APKGEKKPAEKKP---------AEEKKSTVAEKAPAEKKPK 33
A GEK P EKKP +EEKKS EK P EKKPK
Sbjct: 353 AGAGEKSP-EKKPEEKKSDEKKSEEKKSD--EKKPEEKKPK 466
Score = 24.3 bits (51), Expect = 9.4
Identities = 13/20 (65%), Positives = 14/20 (70%)
Frame = +2
Query: 6 EKKPAEKKPAEEKKSTVAEK 25
EKKP EKKP K+STV K
Sbjct: 437 EKKPEEKKP---KESTVVMK 487
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.306 0.125 0.327
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,627,196
Number of Sequences: 28460
Number of extensions: 15655
Number of successful extensions: 221
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 149
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 171
length of query: 148
length of database: 4,897,600
effective HSP length: 82
effective length of query: 66
effective length of database: 2,563,880
effective search space: 169216080
effective search space used: 169216080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 51 (24.3 bits)
Medicago: description of AC139841.13


