

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139747.2 - phase: 0
(1083 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
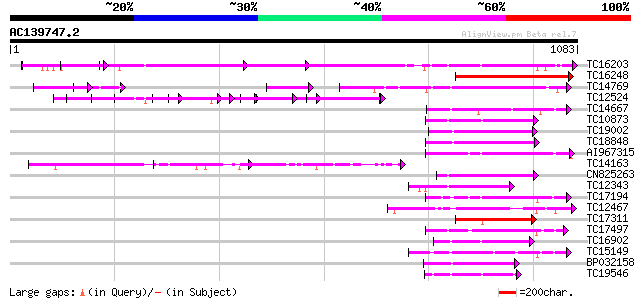
Score E
Sequences producing significant alignments: (bits) Value
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 453 e-128
TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat re... 273 1e-73
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 183 1e-46
TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kina... 165 3e-41
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 157 8e-39
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 157 1e-38
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 153 2e-37
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 150 1e-36
AI967315 149 2e-36
TC14163 140 8e-34
CN825263 140 1e-33
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 139 2e-33
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 139 2e-33
TC12467 UP|CAE02597 (CAE02597) Nod-factor receptor 5, complete 137 7e-33
TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serin... 135 3e-32
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 135 3e-32
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 132 4e-31
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 129 2e-30
BP032158 128 4e-30
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 128 4e-30
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 453 bits (1166), Expect = e-128
Identities = 307/936 (32%), Positives = 480/936 (50%), Gaps = 23/936 (2%)
Frame = +2
Query: 171 RNLR--ELSISYANLTGTIPTSIGNLTLLSHLYLGGNNLYGDIPNELWNLNNLTFLRVEL 228
+NLR L+++ L G +P IG L L +L + NNL +P++L +L +L L +
Sbjct: 323 QNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISH 502
Query: 229 NKFNGSVLAQEIVKLHKIETLDLGGNSLSINGPILQEILKLGNLKYLSFFQCNVRGSIPF 288
N F+G V + ++E LD NS S GP+ +EI+KL LKYL G+IP
Sbjct: 503 NLFSGQFPGNITVGMTELEALDAYDNSFS--GPLPEEIVKLEKLKYLHLAGNYFSGTIPE 676
Query: 289 SIGKLANLSYLNLAHNPISGHLPMEIGKLRKLEYLYI-FDNNLSGSIPVEIGELVKMKEL 347
S + +L +L L N ++G +P + KL+ L+ L++ + N G IP G + ++ L
Sbjct: 677 SYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLL 856
Query: 348 RFNDNNLSGSIPREIGMLRNVVQMDLNNNSLSGEIPPTIGNLSNIQQLSFSLNNLNGKLP 407
+ NL+G IP +G L + + + N+L+G IPP + ++ ++ L S+N+L G++P
Sbjct: 857 EMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIP 1036
Query: 408 MGMNMLLSLENLQIFDNDFIGQLPHNICIGGNLKFLGALNNHFTGRVPKSLKNCSSIIRL 467
+ L +L + F N F G LP I NL+ L N+F+ +P +L +
Sbjct: 1037 ESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYF 1216
Query: 468 RLDQNQLTGNITQDFSVYPNLNYIDLSENNFYGHLSSNWGKCQNLTSFIISHNNISGHIP 527
+ +N LTG I D L +++N F G + G+C++LT +++N + G +P
Sbjct: 1217 DVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVP 1396
Query: 528 PEIGRASNLGILDLSSNHLTGKIPKELSNLSLSKLLISNNHLSGNIPVEISSLDELEILD 587
P + + ++ I +LS+N L G++P +S SL L +SNN +G IP + +L L+ L
Sbjct: 1397 PGVFQLPSVTITELSNNRLNGELPSVISGESLGTLTLSNNLFTGKIPAAMKNLRALQSLS 1576
Query: 588 LAENDLSGFITKQLANLPKVWNLNLSHNKLIGNIPVELGQFKILQSLDLSGNFLNGTIPS 647
L N+ G I + +P + +N+S N L G IP + L ++DLS N L G +P
Sbjct: 1577 LDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEVPK 1756
Query: 648 MLTQLKYLETLNISHNNLSGFIPSSFDQMFSLTSVDISYNQLEGPLPNIRAFSSATIE-V 706
+ L L LN+S N +SG +P M SLT++D+S N G +P F +
Sbjct: 1757 GMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQFLVFNYDKT 1936
Query: 707 LRNNNGLCGNISGLEPCLTPRSKSPDR-KIKKVLLIVLPLVLGTLMLATCFKFLYHLYHT 765
N LC P + S R K +V IV+ + L T +L H+
Sbjct: 1937 FAGNPNLCFPHRASCPSVLYDSLRKTRAKTARVRAIVIGIALATAVLLVA--VTVHVVRK 2110
Query: 766 STIGENQVGGNIIVPQNVFTIWNFDG----KMVYENILEATQDFDDKYLIGVGGQGSVYK 821
+ Q W ++ E+++E + ++ +IG GG G VY+
Sbjct: 2111 RRLHRAQA-------------WKLTAFQRLEIKAEDVVECLK---EENIIGKGGAGIVYR 2242
Query: 822 AELHTGQVVAVKKLHPVSNEENLSPKSFTNEIQALTEIRHRNIVNLYGFCSHSQLSFLVY 881
+ G VA+K+L V + F EI+ L +IRHRNI+ L G+ S+ + L+Y
Sbjct: 2243 GSMPNGTDVAIKRL--VGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLY 2416
Query: 882 EFVEKGSLEKILKDDEEAIAFNWKKRVNVIKDVANALCYMHHDCSPPIVHRDISSKNILL 941
E++ GSL + L + W+ R + + A LCYMHHDCSP I+HRD+ S NILL
Sbjct: 2417 EYMPNGSLGEWLHGAKGG-HLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILL 2593
Query: 942 DSECVAHVSDFGTAKLL-DPNLTSS-TSFACTFGYAAPELAYTTKVTEKCDVYSFGVLAL 999
D++ AHV+DFG AK L DP + S +S A ++GY APE AYT KV EK DVYSFGV+ L
Sbjct: 2594 DADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLL 2773
Query: 1000 EILFGKHP----GDVVPLWTIVTSTL-------DTMPLMDKLDQRLP-RPLNPIVKNLVS 1047
E++ G+ P GD V + V T+ DT ++ +D RL PL ++ ++ +
Sbjct: 2774 ELIIGRKPVGEFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYPLTSVI-HMFN 2950
Query: 1048 IAMIAFTCLTESSQSRPTMEHVAKELAMSKWSRSNS 1083
IAM+ C+ E +RPTM V L S +++
Sbjct: 2951 IAMM---CVKEMGPARPTMREVVHMLTNPPQSNTST 3049
Score = 261 bits (668), Expect = 3e-70
Identities = 188/606 (31%), Positives = 295/606 (48%), Gaps = 56/606 (9%)
Frame = +2
Query: 25 FTTTLSETSQASALLKWKASLDNHSQT--LLSSWSGNNS----CNWLGISCKEDSISVSK 78
+T S S ALLK K S+ L W + S C++ G++C + ++ V
Sbjct: 167 WTVVYSSFSDLDALLKLKESMKGAKAKHHALEDWKFSTSLSAHCSFSGVTC-DQNLRVVA 343
Query: 79 VNLT----------NMGLKGTLESLNFS-------------SLPNIQTLNISHNSLNGSI 115
+N+T +GL LE+L S SL +++ LNISHN +G
Sbjct: 344 LNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQF 523
Query: 116 PSHIGM-LSKLTHLDLSDNLFSGTIPYEITHLISLQTLYLDTNVFSGSIPEEIGELRNLR 174
P +I + +++L LD DN FSG +P EI L L+ L+L N FSG+IPE E ++L
Sbjct: 524 PGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLE 703
Query: 175 ELSISYANLTGTIPTSIGNLTLLSHLYLGGNNLY-------------------------G 209
L ++ +LTG +P S+ L L L+LG +N Y G
Sbjct: 704 FLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTG 883
Query: 210 DIPNELWNLNNLTFLRVELNKFNGSVLAQEIVKLHKIETLDLGGNSLSINGPILQEILKL 269
+IP L NL L L V++N G++ E+ + + +LDL N L+ G I + KL
Sbjct: 884 EIPPSLGNLTKLHSLFVQMNNLTGTI-PPELSSMMSLMSLDLSINDLT--GEIPESFSKL 1054
Query: 270 GNLKYLSFFQCNVRGSIPFSIGKLANLSYLNLAHNPISGHLPMEIGKLRKLEYLYIFDNN 329
NL ++FFQ RGS+P IG L NL L + N S LP +G + Y + N+
Sbjct: 1055KNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNH 1234
Query: 330 LSGSIPVEIGELVKMKELRFNDNNLSGSIPREIGMLRNVVQMDLNNNSLSGEIPPTIGNL 389
L+G IP ++ + ++K DN G IP+ IG R++ ++ + NN L G +PP + L
Sbjct: 1235LTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQL 1414
Query: 390 SNIQQLSFSLNNLNGKLPMGMNMLLSLENLQIFDNDFIGQLPHNICIGGNLKFLGALNNH 449
++ S N LNG+LP ++ G +L L NN
Sbjct: 1415PSVTITELSNNRLNGELP-------------------------SVISGESLGTLTLSNNL 1519
Query: 450 FTGRVPKSLKNCSSIIRLRLDQNQLTGNITQDFSVYPNLNYIDLSENNFYGHLSSNWGKC 509
FTG++P ++KN ++ L LD N+ G I P L +++S NN G + +
Sbjct: 1520FTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHR 1699
Query: 510 QNLTSFIISHNNISGHIPPEIGRASNLGILDLSSNHLTGKIPKELSNL-SLSKLLISNNH 568
+LT+ +S NN++G +P + +L IL+LS N ++G +P E+ + SL+ L +S+N+
Sbjct: 1700ASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNN 1879
Query: 569 LSGNIP 574
+G +P
Sbjct: 1880FTGTVP 1897
Score = 161 bits (407), Expect = 6e-40
Identities = 120/444 (27%), Positives = 217/444 (48%), Gaps = 11/444 (2%)
Frame = +2
Query: 23 SAFTTTLSETSQASALLKWKASLDNH-SQTLLSSWSGNNSCNWLGISCK-------EDSI 74
++F+ L E LK+ N+ S T+ S+S S +LG++ E
Sbjct: 578 NSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLA 757
Query: 75 SVSKVNLTNMGLKGTLES---LNFSSLPNIQTLNISHNSLNGSIPSHIGMLSKLTHLDLS 131
+ + ++G E F S+ N++ L +++ +L G IP +G L+KL L +
Sbjct: 758 KLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQ 937
Query: 132 DNLFSGTIPYEITHLISLQTLYLDTNVFSGSIPEEIGELRNLRELSISYANLTGTIPTSI 191
N +GTIP E++ ++SL +L L N +G IPE +L+NL ++ G++P+ I
Sbjct: 938 MNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFI 1117
Query: 192 GNLTLLSHLYLGGNNLYGDIPNELWNLNNLTFLRVELNKFNGSVLAQEIVKLHKIETLDL 251
G+L L L + NN +P+ L + V N G ++ ++ K +++T +
Sbjct: 1118 GDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTG-LIPPDLCKSGRLKTFII 1294
Query: 252 GGNSLSINGPILQEILKLGNLKYLSFFQCNVRGSIPFSIGKLANLSYLNLAHNPISGHLP 311
N GPI + I + +L + + G +P + +L +++ L++N ++G LP
Sbjct: 1295 TDN--FFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELP 1468
Query: 312 MEIGKLRKLEYLYIFDNNLSGSIPVEIGELVKMKELRFNDNNLSGSIPREIGMLRNVVQM 371
I L L + +N +G IP + L ++ L + N G IP + + + ++
Sbjct: 1469 SVISG-ESLGTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPMLTKV 1645
Query: 372 DLNNNSLSGEIPPTIGNLSNIQQLSFSLNNLNGKLPMGMNMLLSLENLQIFDNDFIGQLP 431
+++ N+L+G IP TI + +++ + S NNL G++P GM L+ L L + N+ G +P
Sbjct: 1646 NISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVP 1825
Query: 432 HNICIGGNLKFLGALNNHFTGRVP 455
I +L L +N+FTG VP
Sbjct: 1826 DEIRFMTSLTTLDLSSNNFTGTVP 1897
Score = 70.1 bits (170), Expect = 2e-12
Identities = 37/92 (40%), Positives = 55/92 (59%)
Frame = +2
Query: 98 LPNIQTLNISHNSLNGSIPSHIGMLSKLTHLDLSDNLFSGTIPYEITHLISLQTLYLDTN 157
+P + +NIS N+L G IP+ I + LT +DLS N +G +P + +L+ L L L N
Sbjct: 1625 IPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRN 1804
Query: 158 VFSGSIPEEIGELRNLRELSISYANLTGTIPT 189
SG +P+EI + +L L +S N TGT+PT
Sbjct: 1805 EISGPVPDEIRFMTSLTTLDLSSNNFTGTVPT 1900
>TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat receptor-like
protein kinase 1, partial (17%)
Length = 866
Score = 273 bits (697), Expect = 1e-73
Identities = 134/229 (58%), Positives = 171/229 (74%), Gaps = 2/229 (0%)
Frame = +2
Query: 851 NEIQALTEIRHRNIVNLYGFCSHSQLSFLVYEFVEKGSLEKILKDDEEAIAFNWKKRVNV 910
+EIQ LTEIRHRN++ L+GFC HS+ SFLVYEF+E GSL+++L D +A AF+W+KRVNV
Sbjct: 2 SEIQTLTEIRHRNVIKLHGFCLHSKFSFLVYEFLEGGSLDQVLNSDTQAAAFHWEKRVNV 181
Query: 911 IKDVANALCYMHHDCSPPIVHRDISSKNILLDSECVAHVSDFGTAKLLDPNLTSSTSFAC 970
+K VANAL YMHHDCSPPI+HRDISSKN+LLD + AHVSDFGTAK L P + T+FA
Sbjct: 182 VKGVANALSYMHHDCSPPIIHRDISSKNVLLDLQYEAHVSDFGTAKFLKPGSHTWTAFAG 361
Query: 971 TFGYAAPELAYTTKVTEKCDVYSFGVLALEILFGKHPGDVVP--LWTIVTSTLDTMPLMD 1028
TFGYAAPELA T +V EKCDVYSFGV ALEI+ G HPGD++ + ++ + L+D
Sbjct: 362 TFGYAAPELAQTMQVNEKCDVYSFGVFALEIIMGMHPGDLISSLMSPSTVPMVNDLLLID 541
Query: 1029 KLDQRLPRPLNPIVKNLVSIAMIAFTCLTESSQSRPTMEHVAKELAMSK 1077
LDQR + PI + ++ IA +A CL ++ SRPTM+ V+K L + K
Sbjct: 542 ILDQRPHQVEKPIDEEVILIASLALACLRKNPHSRPTMDQVSKALVLGK 688
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 183 bits (464), Expect = 1e-46
Identities = 147/475 (30%), Positives = 230/475 (47%), Gaps = 31/475 (6%)
Frame = +3
Query: 630 ILQSLDLSGNFLNGTIPSMLTQLKYLETLNISHNNLSGFIPSSFDQMFSLTSVDISYNQL 689
++ LDLS + L G IPS + ++ LETLNISHN+ G +PS F L SVD+SYN L
Sbjct: 1596 VITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPS-FPLSSLLISVDLSYNDL 1772
Query: 690 EGPLP-------NIRAFSSATIEVLRNNNGLCGNISGLEPCLTPRSKSPDRKIKKVLLIV 742
G LP ++++ E + + N S L R K + + +V++I
Sbjct: 1773 MGKLPESIVKLPHLKSLYFGCNEHMSPEDPANMN-SSLINTDYGRCKGKESRFGQVIVIG 1949
Query: 743 LPLVLGTLMLATCFKFLYHLYHTSTI--GENQVGGNIIVPQNV-FTIWNFDGKMV----- 794
+ G+L++ F L+ + + E G + N+ F++ + D +
Sbjct: 1950 A-ITCGSLLITLAFGVLFVCRYRQKLIPWEGFAGKKYPMETNIIFSLPSKDDFFIKSVSI 2126
Query: 795 ----YENILEATQDFDDKYLIGVGGQGSVYKAELHTGQVVAVKKLHPVSNEENLSPKSFT 850
E I AT+ + K LIG GG GSVY+ L+ GQ VAVK S + + F
Sbjct: 2127 QAFTLEYIEVATERY--KTLIGEGGFGSVYRGTLNDGQEVAVKVRSSTSTQ---GTREFD 2291
Query: 851 NEIQALTEIRHRNIVNLYGFCSHSQLSFLVYEFVEKGSLE-KILKDDEEAIAFNWKKRVN 909
NE+ L+ I+H N+V L G+C+ S LVY F+ GSL+ ++ + + +W R++
Sbjct: 2292 NELNLLSAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLS 2471
Query: 910 VIKDVANALCYMHHDCSPPIVHRDISSKNILLDSECVAHVSDFGTAKLLDPNLTSSTSFA 969
+ A L Y+H ++HRDI S NILLD A V+DFG +K S S
Sbjct: 2472 IALGAARGLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLE 2651
Query: 970 C--TFGYAAPELAYTTKVTEKCDVYSFGVLALEILFGKHPGDVV---PLWTIVTSTLDTM 1024
T GY PE T +++EK DV+SFGV+ LEI+ G+ P ++ W++V +
Sbjct: 2652 VRGTAGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRTEWSLVEWATPYI 2831
Query: 1025 PLMDKLDQRLPRPLNPIVKN------LVSIAMIAFTCLTESSQSRPTMEHVAKEL 1073
++ ++P +K + + +A CL S RP+M + +EL
Sbjct: 2832 -----RGSKVDEIVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIVREL 2981
Score = 71.6 bits (174), Expect = 6e-13
Identities = 45/117 (38%), Positives = 66/117 (55%), Gaps = 4/117 (3%)
Frame = +3
Query: 45 LDNHSQTLLSSWSGNNSC--NWLGISC--KEDSISVSKVNLTNMGLKGTLESLNFSSLPN 100
L N L SWSG+ W GI+C S ++K++L++ LKG + S + + + N
Sbjct: 1494 LQNSGNRALESWSGDPCILLPWKGIACDGSNGSSVITKLDLSSSNLKGLIPS-SIAEMTN 1670
Query: 101 IQTLNISHNSLNGSIPSHIGMLSKLTHLDLSDNLFSGTIPYEITHLISLQTLYLDTN 157
++TLNISHNS +GS+PS + S L +DLS N G +P I L L++LY N
Sbjct: 1671 LETLNISHNSFDGSVPS-FPLSSLLISVDLSYNDLMGKLPESIVKLPHLKSLYFGCN 1838
Score = 53.1 bits (126), Expect = 2e-07
Identities = 36/99 (36%), Positives = 51/99 (51%), Gaps = 1/99 (1%)
Frame = +3
Query: 123 SKLTHLDLSDNLFSGTIPYEITHLISLQTLYLDTNVFSGSIPE-EIGELRNLRELSISYA 181
S +T LDLS + G IP I + +L+TL + N F GS+P + L L + +SY
Sbjct: 1593 SVITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPLSSL--LISVDLSYN 1766
Query: 182 NLTGTIPTSIGNLTLLSHLYLGGNNLYGDIPNELWNLNN 220
+L G +P SI L L LY G N P + N+N+
Sbjct: 1767 DLMGKLPESIVKLPHLKSLYFGCNEHMS--PEDPANMNS 1877
Score = 48.9 bits (115), Expect = 4e-06
Identities = 32/91 (35%), Positives = 48/91 (52%), Gaps = 2/91 (2%)
Frame = +3
Query: 491 IDLSENNFYGHLSSNWGKCQNLTSFIISHNNISGHIPPEIGRASNLGILDLSSNHLTGKI 550
+DLS +N G + S+ + NL + ISHN+ G + P +S L +DLS N L GK+
Sbjct: 1608 LDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSV-PSFPLSSLLISVDLSYNDLMGKL 1784
Query: 551 PKELSNLSLSKLLI--SNNHLSGNIPVEISS 579
P+ + L K L N H+S P ++S
Sbjct: 1785 PESIVKLPHLKSLYFGCNEHMSPEDPANMNS 1877
>TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1,
partial (27%)
Length = 834
Score = 165 bits (418), Expect = 3e-41
Identities = 90/276 (32%), Positives = 144/276 (51%)
Frame = +2
Query: 274 YLSFFQCNVRGSIPFSIGKLANLSYLNLAHNPISGHLPMEIGKLRKLEYLYIFDNNLSGS 333
YL ++ N +G IP IG L L + A+ +SG +P E+GKL+KL+ L++ N LSGS
Sbjct: 8 YLGYYN-NYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGS 184
Query: 334 IPVEIGELVKMKELRFNDNNLSGSIPREIGMLRNVVQMDLNNNSLSGEIPPTIGNLSNIQ 393
+ E+G L +K + ++N LSG +P L+N+ ++L N L G IP +G + ++
Sbjct: 185 LTPELGHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALE 364
Query: 394 QLSFSLNNLNGKLPMGMNMLLSLENLQIFDNDFIGQLPHNICIGGNLKFLGALNNHFTGR 453
L NN G +P + L + + N G LP ++C G L+ L AL N G
Sbjct: 365 VLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGP 544
Query: 454 VPKSLKNCSSIIRLRLDQNQLTGNITQDFSVYPNLNYIDLSENNFYGHLSSNWGKCQNLT 513
+P+SL C S+ R+R+ QN L G+I + P L ++ +N G N+
Sbjct: 545 IPESLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIG 724
Query: 514 SFIISHNNISGHIPPEIGRASNLGILDLSSNHLTGK 549
+S+N +SG +P IG +++ L L N +G+
Sbjct: 725 QITLSNNKLSGPLPSTIGNFTSMQKLLLDGNKFSGR 832
Score = 147 bits (372), Expect = 7e-36
Identities = 100/305 (32%), Positives = 152/305 (49%), Gaps = 7/305 (2%)
Frame = +2
Query: 108 HNSLNGSIPSHIGMLSKLTHLDLSDNLFSGTIPYEITHLISLQTLYLDTNVFSGSIPEEI 167
+N+ G IP IG L++L D + SG IP E+ L L TL+L NV SGS+ E+
Sbjct: 20 YNNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPEL 199
Query: 168 GELRNLRELSISYANLTGTIPTSIGNLTLLSHLYLGGNNLYGDIPNELWNLNNLTFLRVE 227
G L++L+ + +S L+G +P S L L+ L L N L+G IP + + L L++
Sbjct: 200 GHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLW 379
Query: 228 LNKFNGSVLAQEIVKLHKIETLDLGGNSLS-------INGPILQEILKLGNLKYLSFFQC 280
N F GS+ Q + K K+ +DL N L+ +G LQ ++ LGN +
Sbjct: 380 ENNFTGSI-PQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLF------ 538
Query: 281 NVRGSIPFSIGKLANLSYLNLAHNPISGHLPMEIGKLRKLEYLYIFDNNLSGSIPVEIGE 340
G IP S+GK +L+ + + N L+GSIP +
Sbjct: 539 ---GPIPESLGKCESLTRIRMG------------------------QNFLNGSIPKGLFG 637
Query: 341 LVKMKELRFNDNNLSGSIPREIGMLRNVVQMDLNNNSLSGEIPPTIGNLSNIQQLSFSLN 400
L K+ ++ F DN LSG P + N+ Q+ L+NN LSG +P TIGN +++Q+L N
Sbjct: 638 LPKLTQVEFQDNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNFTSMQKLLLDGN 817
Query: 401 NLNGK 405
+G+
Sbjct: 818 KFSGR 832
Score = 141 bits (355), Expect = 6e-34
Identities = 91/273 (33%), Positives = 142/273 (51%)
Frame = +2
Query: 157 NVFSGSIPEEIGELRNLRELSISYANLTGTIPTSIGNLTLLSHLYLGGNNLYGDIPNELW 216
N + G IP EIG L L +Y L+G IP +G L L L+L N L G + EL
Sbjct: 23 NNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELG 202
Query: 217 NLNNLTFLRVELNKFNGSVLAQEIVKLHKIETLDLGGNSLSINGPILQEILKLGNLKYLS 276
+L +L + + N +G V A +L + L+L N L +G I + + ++ L+ L
Sbjct: 203 HLKSLKSMDLSNNMLSGQVPAS-FAELKNLTLLNLFRNRL--HGAIPEFVGEMPALEVLQ 373
Query: 277 FFQCNVRGSIPFSIGKLANLSYLNLAHNPISGHLPMEIGKLRKLEYLYIFDNNLSGSIPV 336
++ N GSIP S+GK L+ ++L+ N ++G LP + +L+ L N L G IP
Sbjct: 374 LWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPE 553
Query: 337 EIGELVKMKELRFNDNNLSGSIPREIGMLRNVVQMDLNNNSLSGEIPPTIGNLSNIQQLS 396
+G+ + +R N L+GSIP+ + L + Q++ +N LSGE P T NI Q++
Sbjct: 554 SLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQIT 733
Query: 397 FSLNNLNGKLPMGMNMLLSLENLQIFDNDFIGQ 429
S N L+G LP + S++ L + N F G+
Sbjct: 734 LSNNKLSGPLPSTIGNFTSMQKLLLDGNKFSGR 832
Score = 137 bits (345), Expect = 9e-33
Identities = 90/294 (30%), Positives = 146/294 (49%), Gaps = 1/294 (0%)
Frame = +2
Query: 303 HNPISGHLPMEIGKLRKLEYLYIFDNNLSGSIPVEIGELVKMKELRFNDNNLSGSIPREI 362
+N G +P EIG L +L LSG IP E+G+L K+ L N LSGS+ E+
Sbjct: 20 YNNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPEL 199
Query: 363 GMLRNVVQMDLNNNSLSGEIPPTIGNLSNIQQLSFSLNNLNGKLPMGMNMLLSLENLQIF 422
G L+++ MDL+NN LSG++P + L N+ L+ N L+G +P
Sbjct: 200 GHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIP--------------- 334
Query: 423 DNDFIGQLPHNICIGGNLKFLGALNNHFTGRVPKSLKNCSSIIRLRLDQNQLTGNITQDF 482
+F+G++P L+ L N+FTG +P+SL + + L N+LTG +
Sbjct: 335 --EFVGEMPA-------LEVLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHM 487
Query: 483 SVYPNLNYIDLSENNFYGHLSSNWGKCQNLTSFIISHNNISGHIPPEIGRASNLGILDLS 542
L + N +G + + GKC++LT + N ++G IP + L ++
Sbjct: 488 CSGNRLQTLIALGNFLFGPIPESLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQ 667
Query: 543 SNHLTGKIPKELS-NLSLSKLLISNNHLSGNIPVEISSLDELEILDLAENDLSG 595
N L+G+ P+ S + ++ ++ +SNN LSG +P I + ++ L L N SG
Sbjct: 668 DNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNFTSMQKLLLDGNKFSG 829
Score = 123 bits (309), Expect = 1e-28
Identities = 86/302 (28%), Positives = 143/302 (46%), Gaps = 25/302 (8%)
Frame = +2
Query: 200 LYLGG-NNLYGDIPNELWNLNNLTFLRVELNKFNGSVLAQEIVKLHKIETLDLGGNSLSI 258
LYLG NN G IP E+ NL L +G + A E+ KL K++TL L N LS
Sbjct: 5 LYLGYYNNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPA-ELGKLQKLDTLFLQVNVLS- 178
Query: 259 NGPILQEILKLGNLKYLSFFQCNVRGSIPFSIGKLANLSYLNLAHNPISGHLPMEIGKLR 318
GS+ +G L +L ++L++N +SG +P +L+
Sbjct: 179 -------------------------GSLTPELGHLKSLKSMDLSNNMLSGQVPASFAELK 283
Query: 319 KLEYLYIFDNNLSGSIPVEIGELVKMKELRFNDNNLSGSIPREIGMLRNVVQMDLNNNSL 378
L L +F N L G+IP +GE+ ++ L+ +NN +GSIP+ +G + +DL++N L
Sbjct: 284 NLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKL 463
Query: 379 SGEIPP------------------------TIGNLSNIQQLSFSLNNLNGKLPMGMNMLL 414
+G +PP ++G ++ ++ N LNG +P G+ L
Sbjct: 464 TGTLPPHMCSGNRLQTLIALGNFLFGPIPESLGKCESLTRIRMGQNFLNGSIPKGLFGLP 643
Query: 415 SLENLQIFDNDFIGQLPHNICIGGNLKFLGALNNHFTGRVPKSLKNCSSIIRLRLDQNQL 474
L ++ DN G+ P + N+ + NN +G +P ++ N +S+ +L LD N+
Sbjct: 644 KLTQVEFQDNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNFTSMQKLLLDGNKF 823
Query: 475 TG 476
+G
Sbjct: 824 SG 829
Score = 122 bits (307), Expect = 2e-28
Identities = 81/247 (32%), Positives = 120/247 (47%), Gaps = 3/247 (1%)
Frame = +2
Query: 472 NQLTGNITQDFSVYPNLNYIDLSENNFYGHLSSNWGKCQNLTSFIISHNNISGHIPPEIG 531
N G I + L D + G + + GK Q L + + N +SG + PE+G
Sbjct: 23 NNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELG 202
Query: 532 RASNLGILDLSSNHLTGKIPK---ELSNLSLSKLLISNNHLSGNIPVEISSLDELEILDL 588
+L +DLS+N L+G++P EL NL+L L N L G IP + + LE+L L
Sbjct: 203 HLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLF--RNRLHGAIPEFVGEMPALEVLQL 376
Query: 589 AENDLSGFITKQLANLPKVWNLNLSHNKLIGNIPVELGQFKILQSLDLSGNFLNGTIPSM 648
EN+ +G I + L K+ ++LS NKL G +P + LQ+L GNFL G IP
Sbjct: 377 WENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPES 556
Query: 649 LTQLKYLETLNISHNNLSGFIPSSFDQMFSLTSVDISYNQLEGPLPNIRAFSSATIEVLR 708
L + + L + + N L+G IP + LT V+ N L G P + S ++
Sbjct: 557 LGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITL 736
Query: 709 NNNGLCG 715
+NN L G
Sbjct: 737 SNNKLSG 757
Score = 122 bits (305), Expect = 4e-28
Identities = 79/275 (28%), Positives = 136/275 (48%), Gaps = 1/275 (0%)
Frame = +2
Query: 442 FLGALNNHFTGRVPKSLKNCSSIIRLRLDQNQLTGNITQDFSVYPNLNYIDLSENNFYGH 501
+LG NN + G +P + N + ++R L+G I + L+ + L N G
Sbjct: 8 YLGYYNN-YQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGS 184
Query: 502 LSSNWGKCQNLTSFIISHNNISGHIPPEIGRASNLGILDLSSNHLTGKIPKELSNL-SLS 560
L+ G ++L S +S+N +SG +P NL +L+L N L G IP+ + + +L
Sbjct: 185 LTPELGHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALE 364
Query: 561 KLLISNNHLSGNIPVEISSLDELEILDLAENDLSGFITKQLANLPKVWNLNLSHNKLIGN 620
L + N+ +G+IP + +L ++DL+ N L+G + + + ++ L N L G
Sbjct: 365 VLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGP 544
Query: 621 IPVELGQFKILQSLDLSGNFLNGTIPSMLTQLKYLETLNISHNNLSGFIPSSFDQMFSLT 680
IP LG+ + L + + NFLNG+IP L L L + N LSG P + ++
Sbjct: 545 IPESLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIG 724
Query: 681 SVDISYNQLEGPLPNIRAFSSATIEVLRNNNGLCG 715
+ +S N+L GPLP+ ++ ++L + N G
Sbjct: 725 QITLSNNKLSGPLPSTIGNFTSMQKLLLDGNKFSG 829
Score = 110 bits (274), Expect = 2e-24
Identities = 76/248 (30%), Positives = 119/248 (47%)
Frame = +2
Query: 85 GLKGTLESLNFSSLPNIQTLNISHNSLNGSIPSHIGMLSKLTHLDLSDNLFSGTIPYEIT 144
GL G + + L + TL + N L+GS+ +G L L +DLS+N+ SG +P
Sbjct: 98 GLSGEIPA-ELGKLQKLDTLFLQVNVLSGSLTPELGHLKSLKSMDLSNNMLSGQVPASFA 274
Query: 145 HLISLQTLYLDTNVFSGSIPEEIGELRNLRELSISYANLTGTIPTSIGNLTLLSHLYLGG 204
L +L L L N G+IPE +GE+ L L + N TG+IP S+G L+ + L
Sbjct: 275 ELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWENNFTGSIPQSLGKNGKLTLVDLSS 454
Query: 205 NNLYGDIPNELWNLNNLTFLRVELNKFNGSVLAQEIVKLHKIETLDLGGNSLSINGPILQ 264
N L G +P + + N L L + L F + + + K + + +G N L NG I +
Sbjct: 455 NKLTGTLPPHMCSGNRLQTL-IALGNFLFGPIPESLGKCESLTRIRMGQNFL--NGSIPK 625
Query: 265 EILKLGNLKYLSFFQCNVRGSIPFSIGKLANLSYLNLAHNPISGHLPMEIGKLRKLEYLY 324
+ L L + F + G P + N+ + L++N +SG LP IG ++ L
Sbjct: 626 GLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNFTSMQKLL 805
Query: 325 IFDNNLSG 332
+ N SG
Sbjct: 806 LDGNKFSG 829
Score = 85.5 bits (210), Expect = 4e-17
Identities = 54/151 (35%), Positives = 77/151 (50%)
Frame = +2
Query: 567 NHLSGNIPVEISSLDELEILDLAENDLSGFITKQLANLPKVWNLNLSHNKLIGNIPVELG 626
N+ G IP EI +L +L D A LSG I +L L K+ L L N L G++ ELG
Sbjct: 23 NNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELG 202
Query: 627 QFKILQSLDLSGNFLNGTIPSMLTQLKYLETLNISHNNLSGFIPSSFDQMFSLTSVDISY 686
K L+S+DLS N L+G +P+ +LK L LN+ N L G IP +M +L + +
Sbjct: 203 HLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWE 382
Query: 687 NQLEGPLPNIRAFSSATIEVLRNNNGLCGNI 717
N G +P + V ++N L G +
Sbjct: 383 NNFTGSIPQSLGKNGKLTLVDLSSNKLTGTL 475
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 157 bits (397), Expect = 8e-39
Identities = 103/293 (35%), Positives = 149/293 (50%), Gaps = 15/293 (5%)
Frame = +2
Query: 796 ENILEATQDFDDKYLIGVGGQGSVYKAELHTGQVVAVKKLHPVSNEENLSPKSFTNEIQA 855
+ + E T +F K LIG G G VY A L+ G VAVKKL S E + F ++
Sbjct: 332 DELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPE--TNNEFLTQVSM 505
Query: 856 LTEIRHRNIVNLYGFCSHSQLSFLVYEFVEKGSLEKIL------KDDEEAIAFNWKKRVN 909
++ +++ N V L+G+C L L YEF GSL IL + + +W +RV
Sbjct: 506 VSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRVR 685
Query: 910 VIKDVANALCYMHHDCSPPIVHRDISSKNILLDSECVAHVSDFGTAKLLDPNLTS---ST 966
+ D A L Y+H P I+HRDI S N+L+ + A ++DF + P++ + ST
Sbjct: 686 IAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQA-PDMAARLHST 862
Query: 967 SFACTFGYAAPELAYTTKVTEKCDVYSFGVLALEILFGKHPGD-VVP-----LWTIVTST 1020
TFGY APE A T ++T+K DVYSFGV+ LE+L G+ P D +P L T T
Sbjct: 863 RVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATPR 1042
Query: 1021 LDTMPLMDKLDQRLPRPLNPIVKNLVSIAMIAFTCLTESSQSRPTMEHVAKEL 1073
L + +D +L P K + +A +A C+ ++ RP M V K L
Sbjct: 1043 LSEDKVKQCVDPKLKGEYPP--KGVAKLAAVAALCVQYEAEFRPNMSIVVKAL 1195
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 157 bits (396), Expect = 1e-38
Identities = 91/220 (41%), Positives = 129/220 (58%), Gaps = 5/220 (2%)
Frame = +1
Query: 795 YENILEATQDFDDKYLIGVGGQGSVYKAELHTGQVVAVKKLHPVSNEENLSPKSFTNEIQ 854
+ + +AT++F + LIG GG G VYK L TG+ VAVK+L S++ + F E+
Sbjct: 421 FRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQL---SHDGRQGFQEFVMEVL 591
Query: 855 ALTEIRHRNIVNLYGFCSHSQLSFLVYEFVEKGSLEKILKD---DEEAIAFNWKKRVNVI 911
L+ + H N+V L G+C+ LVYE++ GSLE L + D+E + NW R+ V
Sbjct: 592 MLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPL--NWSTRMKVA 765
Query: 912 KDVANALCYMHHDCSPPIVHRDISSKNILLDSECVAHVSDFGTAKL--LDPNLTSSTSFA 969
A L Y+H PP+++RD+ S NILLD+E +SDFG AKL + N ST
Sbjct: 766 VGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVM 945
Query: 970 CTFGYAAPELAYTTKVTEKCDVYSFGVLALEILFGKHPGD 1009
T+GY APE A + K+T K D+YSFGV+ LE+L G+ D
Sbjct: 946 GTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAID 1065
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 153 bits (386), Expect = 2e-37
Identities = 82/209 (39%), Positives = 122/209 (58%), Gaps = 2/209 (0%)
Frame = +1
Query: 801 ATQDFDDKYLIGVGGQGSVYKAELHTGQVVAVKKLHPVSNEENLSPKSFTNEIQALTEIR 860
AT +F+ +G GG GSVY +L G +AVK+L SN+ ++ F E++ L +R
Sbjct: 52 ATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADME---FAVEVEILARVR 222
Query: 861 HRNIVNLYGFCSHSQLSFLVYEFVEKGSLEKILKDDEEA-IAFNWKKRVNVIKDVANALC 919
H+N+++L G+C+ Q +VY+++ SL L + +W +R+N+ A +
Sbjct: 223 HKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIGSAEGIV 402
Query: 920 YMHHDCSPPIVHRDISSKNILLDSECVAHVSDFGTAKLLDPNLTS-STSFACTFGYAAPE 978
Y+HH +P I+HRDI + N+LLDS+ A V+DFG AKL+ T +T T GY APE
Sbjct: 403 YLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVTTRVKGTLGYLAPE 582
Query: 979 LAYTTKVTEKCDVYSFGVLALEILFGKHP 1007
A K E CDV+SFG+L LE+ GK P
Sbjct: 583 YAMLGKANECCDVFSFGILLLELASGKKP 669
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 150 bits (378), Expect = 1e-36
Identities = 81/220 (36%), Positives = 126/220 (56%), Gaps = 2/220 (0%)
Frame = +2
Query: 795 YENILEATQDFDDKYLIGVGGQGSVYKAELHTGQVVAVKKLHPVSNEENLSPKSFTNEIQ 854
Y+ + AT F D +G GG GSVY G +AVKKL ++++ + F E++
Sbjct: 272 YKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEME---FAVEVE 442
Query: 855 ALTEIRHRNIVNLYGFCSHSQLSFLVYEFVEKGSLEKILKDDEEA-IAFNWKKRVNVIKD 913
L +RH+N++ L G+C +VY+++ SL L + NW+KR+ +
Sbjct: 443 VLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIAIG 622
Query: 914 VANALCYMHHDCSPPIVHRDISSKNILLDSECVAHVSDFGTAKLLDPNLT-SSTSFACTF 972
A + Y+HH+ +P I+HRDI + N+LL+S+ V+DFG AKL+ ++ +T T
Sbjct: 623 SAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSHMTTRVKGTL 802
Query: 973 GYAAPELAYTTKVTEKCDVYSFGVLALEILFGKHPGDVVP 1012
GY APE A KV+E CDVYSFG+L LE++ G+ P + +P
Sbjct: 803 GYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLP 922
>AI967315
Length = 1308
Score = 149 bits (376), Expect = 2e-36
Identities = 99/293 (33%), Positives = 141/293 (47%), Gaps = 9/293 (3%)
Frame = +1
Query: 795 YENILEATQDFDDKYLIGVGGQGSVYKAELHTGQVVAVKKLHPVSNEENLSPKSFTNEIQ 854
YE + AT F + ++G GG VYK L +G +AVK+L +E K F EI
Sbjct: 118 YEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDER-KEKEFLTEIG 294
Query: 855 ALTEIRHRNIVNLYGFCSHSQLSFLVYEFVEKGSLEKILKDDEEAIAFNWKKRVNVIKDV 914
+ + H N++ L G C + L +LV+E GS+ ++ D++ A +WK R ++
Sbjct: 295 TIGHVCHSNVMPLLGCCIDNGL-YLVFELSTVGSVASLIHDEKMA-PLDWKTRYKIVLGT 468
Query: 915 ANALCYMHHDCSPPIVHRDISSKNILLDSECVAHVSDFGTAKLLDPNLT--SSTSFACTF 972
A L Y+H C I+HRDI + NILL + +SDFG AK L T S TF
Sbjct: 469 ARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPIEGTF 648
Query: 973 GYAAPELAYTTKVTEKCDVYSFGVLALEILFGKHP--GDVVPLWTIVTSTLDTMPLMDKL 1030
G+ APE V EK DV++FGV LE++ G+ P G L T L + +
Sbjct: 649 GHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVDGSHQSLHTWAKPILSKWEIEKLV 828
Query: 1031 DQRLPRPLNPIVKNLVSIAMIAFTCLTESSQSRPTMEHVAK-----ELAMSKW 1078
D RL + V +A A C+ SS RPTM V + E+ +W
Sbjct: 829 DPRLEGCYD--VTQFNRVAFAASLCIRASSTWRPTMSEVLEVMEEGEMDKERW 981
>TC14163
Length = 1712
Score = 140 bits (354), Expect = 8e-34
Identities = 142/493 (28%), Positives = 222/493 (44%), Gaps = 12/493 (2%)
Frame = +1
Query: 276 SFFQCNVRGSIPFSIGKLANLS---YLNLAHNPISGHLPMEIGKLRKLEYLYIFDNNLSG 332
SFF G+I S+ K+ NL LNL + ISG P + KL KL+++YI +N LSG
Sbjct: 370 SFFS----GTISPSLSKIKNLDGFYLLNLKN--ISGPFPGFLFKLPKLQFIYIENNQLSG 531
Query: 333 SIPVEIGELVKMKELRFNDNNLSGSIPREIGMLRNVVQMDLNNNSLSGEIPPTIGNLSNI 392
IP IG L ++ L N +G+IP +G L ++ Q+ L NNSL+G IP TI L N+
Sbjct: 532 RIPENIGNLTRLDVLSLTGNRFTGTIPSSVGGLTHLTQLQLGNNSLTGTIPATIARLKNL 711
Query: 393 QQLSFSLNNLNGKLPMGMNMLLSLENLQIFDNDFIGQLPHNICIGGNLKFLGALNNHFTG 452
LS N +G +P DF +L L N F+G
Sbjct: 712 TYLSLEGNQFSGAIP-----------------DFFSSFT-------DLGILRLSRNKFSG 819
Query: 453 RVPKSLKNCSSIIR-LRLDQNQLTGNITQDFSVYPNLNYIDLSENNFYGHLSSNWGKCQN 511
++P S+ + +R L L NQL+G I + L+ +DLS N F G + +++
Sbjct: 820 KIPASISTLAPKLRYLELGHNQLSGKIPDFLGKFRALDTLDLSSNRFSGTVPASFKNLTK 999
Query: 512 LTSFIISHNNISGHIPPEIGRASNLGILDLSSN--HLTGKIPKELSNLSLSKLLISNNHL 569
+ + +++N + P + LDLS+N HL +IPK +++ S ++ S
Sbjct: 1000IFNLNLANNLLVDPFPEM--NVKGIESLDLSNNMFHL-NQIPKWVTS---SPIIFSLKLA 1161
Query: 570 SGNIPVEISSLDELE-----ILDLAENDLSGFITKQLANLPKVWNLNLSHNKLIGNIPVE 624
I +++ E +DL+ N++SG + + + S NKL E
Sbjct: 1162RCGIKMKLDDWKPAETYFYDFIDLSGNEISGSAVGLVNSTEYLVGFWGSGNKL--KFDFE 1335
Query: 625 LGQF-KILQSLDLSGNFLNGTIPSMLTQLKYLETLNISHNNLSGFIPSSFDQMFSLTSVD 683
+F + + LDLS N + G +P + LE LN+S+N+L G IP +
Sbjct: 1336RLKFGERFKYLDLSHNLVFGKVPKSVAG---LEKLNVSYNHLCGEIPKT----------- 1473
Query: 684 ISYNQLEGPLPNIRAFSSATIEVLRNNNGLCGNISGLEPCLTPRSKSPDRKIKKVLLIVL 743
+ +SA + N+ LCG + L+PC S S +L
Sbjct: 1474-------------KFPASAFV----GNDCLCG--APLQPCKKA*SSS-----------LL 1563
Query: 744 PLVLGTLMLATCF 756
PL++ L+L CF
Sbjct: 1564PLLVLVLLLPLCF 1602
Score = 131 bits (329), Expect = 7e-31
Identities = 122/468 (26%), Positives = 199/468 (42%), Gaps = 38/468 (8%)
Frame = +1
Query: 36 SALLKWKASLDNHSQTLLSSW-SGNNSCNWLGISCKEDSISVSKVNLTNMG------LKG 88
+ L+ +K+ + + +L SW G + C W G++C D V+ + L+ G
Sbjct: 205 AGLMGFKSGIKSDPSGILKSWIPGTDCCTWQGVTCLFDDKRVTSLYLSGNPENPKSFFSG 384
Query: 89 TLESLNFSSLPNIQTLNISH-NSLNGSIPSHIGMLSKLTHLDLSDNLFSGTIPYEITHLI 147
T+ S + S + N+ + + +++G P + L KL + + +N SG IP I +L
Sbjct: 385 TI-SPSLSKIKNLDGFYLLNLKNISGPFPGFLFKLPKLQFIYIENNQLSGRIPENIGNLT 561
Query: 148 SLQTLYLDTNVFSGSIPEEIGELRNLRELSISYANLTGTIPTSIGNLTLLSHLYLGGNNL 207
L L L N F+G+IP +G L +L +L + +LTGTIP +I L L++L L GN
Sbjct: 562 RLDVLSLTGNRFTGTIPSSVGGLTHLTQLQLGNNSLTGTIPATIARLKNLTYLSLEGNQF 741
Query: 208 YGDIPNELWNLNNLTFLRVELNKFNGSVLAQEIVKLHKIETLDLGGNSLSINGPILQEIL 267
G IP+ + +L LR+ NKF+G + A K+ L+LG N LS
Sbjct: 742 SGAIPDFFSSFTDLGILRLSRNKFSGKIPASISTLAPKLRYLELGHNQLS---------- 891
Query: 268 KLGNLKYLSFFQCNVRGSIPFSIGKLANLSYLNLAHNPISGHLPMEIGKLRKLEYLYIFD 327
G IP +GK L L+L+ N SG +P L K+ L + +
Sbjct: 892 ----------------GKIPDFLGKFRALDTLDLSSNRFSGTVPASFKNLTKIFNLNLAN 1023
Query: 328 NNLSGSIP---VEIGELVKMKELRFNDNNL-----SGSIPREIGMLRNVVQM-------- 371
N L P V+ E + + F+ N + S I + + R ++M
Sbjct: 1024NLLVDPFPEMNVKGIESLDLSNNMFHLNQIPKWVTSSPIIFSLKLARCGIKMKLDDWKPA 1203
Query: 372 --------DLNNNSLSGEIPPTIGNLSNIQQLSFSLNNLNGKLPMGMNMLLSLENLQIFD 423
DL+ N +SG +G +++ + L G G + E L+ +
Sbjct: 1204ETYFYDFIDLSGNEISGS---AVGLVNSTEYLV-------GFWGSGNKLKFDFERLKFGE 1353
Query: 424 NDFIGQLPHNICIG------GNLKFLGALNNHFTGRVPKSLKNCSSII 465
L HN+ G L+ L NH G +PK+ S+ +
Sbjct: 1354RFKYLDLSHNLVFGKVPKSVAGLEKLNVSYNHLCGEIPKTKFPASAFV 1497
>CN825263
Length = 663
Score = 140 bits (352), Expect = 1e-33
Identities = 81/199 (40%), Positives = 110/199 (54%), Gaps = 3/199 (1%)
Frame = +1
Query: 815 GQGSVYKAELHTGQVVAVKKLHPVSNEENLSPKSFTNEIQALTEIRHRNIVNLYGFCSHS 874
G G VYK L+ G+ VAVK L ++ + F E++ L+ + HRN+V L G C
Sbjct: 1 GFGLVYKGILNDGRDVAVKIL---KRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEK 171
Query: 875 QLSFLVYEFVEKGSLEKILKD-DEEAIAFNWKKRVNVIKDVANALCYMHHDCSPPIVHRD 933
Q L+YE V GS+E L D+E +W R+ + A L Y+H D +P ++HRD
Sbjct: 172 QTRCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRD 351
Query: 934 ISSKNILLDSECVAHVSDFGTAK--LLDPNLTSSTSFACTFGYAAPELAYTTKVTEKCDV 991
S NILL+ + VSDFG A+ L + N ST TFGY APE A T + K DV
Sbjct: 352 FKSSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDV 531
Query: 992 YSFGVLALEILFGKHPGDV 1010
YS+GV+ LE+L G P D+
Sbjct: 532 YSYGVVLLELLTGTKPVDL 588
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 139 bits (351), Expect = 2e-33
Identities = 78/214 (36%), Positives = 118/214 (54%), Gaps = 13/214 (6%)
Frame = +3
Query: 763 YHTSTIGENQVGGNIIVP---------QNVFTIWNFDGK----MVYENILEATQDFDDKY 809
+H+ N N+I P QN I N K YE ++ AT++F
Sbjct: 78 FHSFMTKSNTFFQNLIKPFKLGSSSEAQNEDDIQNIAAKEHKTFSYETLVAATKNFHAVN 257
Query: 810 LIGVGGQGSVYKAELHTGQVVAVKKLHPVSNEENLSPKSFTNEIQALTEIRHRNIVNLYG 869
+G GG G V+K +L+ G+ +AVKKL S N F NE + LT ++HRN+V+L+G
Sbjct: 258 KLGEGGFGPVFKGKLNDGREIAVKKL---SRRSNQGRTQFINEAKLLTRVQHRNVVSLFG 428
Query: 870 FCSHSQLSFLVYEFVEKGSLEKILKDDEEAIAFNWKKRVNVIKDVANALCYMHHDCSPPI 929
+C+H LVYE+V + SL+K+L ++ +WK+R ++I VA L Y+H D I
Sbjct: 429 YCAHGSEKLLVYEYVPRESLDKLLFRSQKKEQLDWKRRFDIISGVARGLLYLHEDSHDCI 608
Query: 930 VHRDISSKNILLDSECVAHVSDFGTAKLLDPNLT 963
+HRDI + NILLD + V ++DFG A++ + T
Sbjct: 609 IHRDIKAANILLDEKWVPKIADFGLARIFPEDQT 710
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 139 bits (350), Expect = 2e-33
Identities = 96/292 (32%), Positives = 144/292 (48%), Gaps = 13/292 (4%)
Frame = +1
Query: 795 YENILEATQDFDDKYLIGVGGQGSVYKAELHTGQVVAVKKLHPVSNEENLSPKSFTNEIQ 854
Y+ + +AT +F IG GG G+VY AEL G+ A+KK+ ++ E F E++
Sbjct: 1057 YQELAKATNNFSLDNKIGQGGFGAVYYAELR-GKKTAIKKMDVQASTE------FLCELK 1215
Query: 855 ALTEIRHRNIVNLYGFCSHSQLSFLVYEFVEKGSLEKILKDDEEAIAFNWKKRVNVIKDV 914
LT + H N+V L G+C L FLVYE ++ G+L + L + W RV + D
Sbjct: 1216 VLTHVHHLNLVRLIGYCVEGSL-FLVYEHIDNGNLGQYLHGSGKE-PLPWSSRVQIALDA 1389
Query: 915 ANALCYMHHDCSPPIVHRDISSKNILLDSECVAHVSDFGTAKLLD-PNLTSSTSFACTFG 973
A L Y+H P +HRD+ S NIL+D V+DFG KL++ N T T TFG
Sbjct: 1390 ARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNSTLQTRLVGTFG 1569
Query: 974 YAAPELAYTTKVTEKCDVYSFGVLALEILFGKH-----------PGDVVPLWTIVTSTLD 1022
Y PE A ++ K DVY+FGV+ E++ K+ +V L+ + D
Sbjct: 1570 YMPPEYAQYGDISPKIDVYAFGVVLFELISAKNAVLKTGELVAESKGLVALFEEALNKSD 1749
Query: 1023 TMPLMDKL-DQRLPRPLNPIVKNLVSIAMIAFTCLTESSQSRPTMEHVAKEL 1073
+ KL D RL N + +++ IA + C ++ RP+M + L
Sbjct: 1750 PCDALRKLVDPRLGE--NYPIDSVLKIAQLGRACTRDNPLLRPSMRSLVVAL 1899
>TC12467 UP|CAE02597 (CAE02597) Nod-factor receptor 5, complete
Length = 2292
Score = 137 bits (346), Expect = 7e-33
Identities = 118/389 (30%), Positives = 183/389 (46%), Gaps = 28/389 (7%)
Frame = +3
Query: 722 PCLTPRSKSPD--------RKIKKVLLIVLPLVLG-TLMLATCFKFLYHLYHTSTIGENQ 772
P L P ++ P+ RK LL++L + LG TL+ A L ++Y N+
Sbjct: 798 PILIPVTQLPELTQPSSNGRKSSIHLLVILGITLGCTLLTAVLTGTLVYVYCRRKKALNR 977
Query: 773 VGGNIIVPQNVFT-IWNFDGKM-VYE--NILEATQDFDDKYLIGVGGQGSVYKAELHTGQ 828
+ + + + + K VYE I+EAT+DF D+ +G SVYKA + G+
Sbjct: 978 TASSAETADKLLSGVSGYVSKPNVYEIDEIMEATKDFSDECKVGE----SVYKANIE-GR 1142
Query: 829 VVAVKKLHPVSNEENLSPKSFTNEIQALTEIRHRNIVNLYGFCS-HSQLSFLVYEFVEKG 887
VVAVKK+ E E++ L ++ H N+V L G S + FLVYE+ E G
Sbjct: 1143 VVAVKKIKEGGANE---------ELKILQKVNHGNLVKLMGVSSGYDGNCFLVYEYAENG 1295
Query: 888 SLEKIL--KDDEEAIAFNWKKRVNVIKDVANALCYMHHDCSPPIVHRDISSKNILLDSEC 945
SL + L K + W +R+++ DVA L YMH P I+HRDI++ NILLDS
Sbjct: 1296 SLAEWLFSKSSGTPNSLTWSQRISIAVDVAVGLQYMHEHTYPRIIHRDITTSNILLDSNF 1475
Query: 946 VAHVSDFGTAKLLDPNLTSSTSFACTFGYAAPELAYTTKVTEKCDVYSFGVLALEILFG- 1004
A +++F A+ T + K DV++FGVL +E+L G
Sbjct: 1476 KAKIANFAMAR-----------------------TSTNPMMPKIDVFAFGVLLIELLTGR 1586
Query: 1005 -----KHPGDVVPLWTIVTSTLDTMPLMDKLDQRLPRPLNP------IVKNLVSIAMIAF 1053
K G+VV LW + D + + ++R+ + ++P + N +S+A +A
Sbjct: 1587 KAMTTKENGEVVMLWKDMWEIFD---IEENREERIRKWMDPNLESFYHIDNALSLASLAV 1757
Query: 1054 TCLTESSQSRPTMEHVAKELAMSKWSRSN 1082
C + S SRP+M + L+ SN
Sbjct: 1758 NCTADKSLSRPSMAEIVLSLSFLTQQSSN 1844
>TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serine-threonine
kinase 3, partial (7%)
Length = 606
Score = 135 bits (341), Expect = 3e-32
Identities = 74/165 (44%), Positives = 101/165 (60%), Gaps = 11/165 (6%)
Frame = +3
Query: 852 EIQALTEIRHRNIVNLYGFCSHSQLSFLVYEFVEKGSLEKIL--KDDEEAIA-------F 902
E++ L+ IRH NIV L S + LVY+++E SL++ L K ++
Sbjct: 3 EVEILSNIRHNNIVKLQCCISSEESLLLVYDYLENLSLDRWLHKKSKPPTVSGSVQNNII 182
Query: 903 NWKKRVNVIKDVANALCYMHHDCSPPIVHRDISSKNILLDSECVAHVSDFGTA--KLLDP 960
+W +R+++ VA LCYMHHDCSPP+VHRD+ NILLDS+ A V+DFG A +
Sbjct: 183 DWPRRLHIAIGVAQGLCYMHHDCSPPVVHRDLKPSNILLDSQFNAKVADFGLAMMSVKPE 362
Query: 961 NLTSSTSFACTFGYAAPELAYTTKVTEKCDVYSFGVLALEILFGK 1005
L + ++ A TFGY APE A T +V EK DVYSFGV+ LE+ GK
Sbjct: 363 ELATMSAVAGTFGYIAPEYALTIRVNEKIDVYSFGVILLELTTGK 497
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 135 bits (340), Expect = 3e-32
Identities = 92/284 (32%), Positives = 135/284 (47%), Gaps = 12/284 (4%)
Frame = +1
Query: 795 YENILEATQDFDDKYLIGVGGQGSVYKAELHTGQVVAVKKLHPVSNEENLSPKSFTNEIQ 854
+ I AT F +G GG G VYK L GQ +AVK+L SN + F NEI+
Sbjct: 1642 FSTISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRL---SNTSGQGMEEFKNEIK 1812
Query: 855 ALTEIRHRNIVNLYGFCSHSQLSFLVYEFVEKGSLEKILKDDEEAIAFNWKKRVNVIKDV 914
+ ++HRN+V L+G H E KIL D + +W KR+ +I +
Sbjct: 1813 LIARLQHRNLVKLFGCSVHQD------ENSHANKKMKILLDSTRSKLVDWNKRLQIIDGI 1974
Query: 915 ANALCYMHHDCSPPIVHRDISSKNILLDSECVAHVSDFGTAKLL--DPNLTSSTSFACTF 972
A L Y+H D I+HRD+ + NILLD E +SDFG A++ D + T+
Sbjct: 1975 ARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMGTY 2154
Query: 973 GYAAPELAYTTKVTEKCDVYSFGVLALEILFGK----------HPGDVVPLWTIVTSTLD 1022
GY PE A + K DV+SFGV+ LEI+ GK H + W + ++
Sbjct: 2155 GYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLLSHAWRL---WIE 2325
Query: 1023 TMPLMDKLDQRLPRPLNPIVKNLVSIAMIAFTCLTESSQSRPTM 1066
PL + +D+ L P+ P ++ +A C+ ++RP M
Sbjct: 2326 ERPL-ELVDELLDDPVIP--TEILRYIHVALLCVQRRPENRPDM 2448
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like protein,
partial (46%)
Length = 941
Score = 132 bits (331), Expect = 4e-31
Identities = 78/196 (39%), Positives = 114/196 (57%), Gaps = 3/196 (1%)
Frame = +3
Query: 810 LIGVGGQGSVYKAELHTGQVVAVKKLHPVSNEENLSPKSFTNEIQALTEIRHRNIVNLYG 869
L+GVGG G VY E+ G VA+K+ +P+S + F EI+ L+++RHR++V+L G
Sbjct: 12 LLGVGGFGKVYYGEVDGGTKVAIKRGNPLSEQ---GVHEFQTEIEMLSKLRHRHLVSLIG 182
Query: 870 FCSHSQLSFLVYEFVEKGSLEKILKDDEEAIAFNWKKRVNVIKDVANALCYMHHDCSPPI 929
+C + LVY+ + G+L + L ++ WK+R+ + A L Y+H I
Sbjct: 183 YCEENTEMILVYDHMAYGTLREHLYKTQKP-PLPWKQRLEICIGAARGLHYLHTGAKYTI 359
Query: 930 VHRDISSKNILLDSECVAHVSDFGTAK---LLDPNLTSSTSFACTFGYAAPELAYTTKVT 986
+HRD+ + NILLD + VA VSDFG +K LD N ST +FGY PE ++T
Sbjct: 360 IHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLD-NTHVSTVVKGSFGYLDPEYFRRQQLT 536
Query: 987 EKCDVYSFGVLALEIL 1002
+K DVYSFGV+ EIL
Sbjct: 537 DKSDVYSFGVVLFEIL 584
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 129 bits (325), Expect = 2e-30
Identities = 99/321 (30%), Positives = 155/321 (47%), Gaps = 10/321 (3%)
Frame = +3
Query: 763 YHTSTIGENQVGGNIIVPQNVFTIWNFDGKMVYENILEATQDFDDKYLIGVGGQGSVYKA 822
Y + N+V N + + + N E++L A+ + ++G G G+ YKA
Sbjct: 165 YSAAAATINKVEANGVGAKKLVFFGNSARGFDLEDLLRASAE-----VLGKGTFGTAYKA 329
Query: 823 ELHTGQVVAVKKLHPVSNEENLSPKSFTNEIQALTEIRHRNIVNLYGFCSHSQLSFLVYE 882
L TG VVAVK+L V+ +S K F ++I+ + + H+++V L + LVY+
Sbjct: 330 VLETGLVVAVKRLKDVT----ISEKEFKDKIETVGAMDHQSLVPLRAYYFSRDEKLLVYD 497
Query: 883 FVEKGSLEKILKDDEEA--IAFNWKKRVNVIKDVANALCYMHHDCSPPIVHRDISSKNIL 940
++ GSL +L ++ A NW+ R + A + Y+H P + H +I + NIL
Sbjct: 498 YMPMGSLSALLHGNKGAGRTPLNWEIRSGIALGAARGIEYLHSQ-GPNVSHGNIKASNIL 674
Query: 941 LDSECVAHVSDFGTAKLLDPNLTSSTSFACTFGYAAPELAYTTKVTEKCDVYSFGVLALE 1000
L A VSDFG A L+ P+ T + GY APE+ KV++K DVYSFGVL LE
Sbjct: 675 LTKSYEAKVSDFGLAHLVGPSSTPNR----VAGYRAPEVTDPRKVSQKADVYSFGVLLLE 842
Query: 1001 ILFGKHP--------GDVVPLWTIVTSTLDTMPLMDKLDQRLPRPLNPIVKNLVSIAMIA 1052
+L GK P G +P W V S + + D L R N + + +V + +A
Sbjct: 843 LLTGKAPTHALLNEEGVDLPRW--VQSLVREEWTSEVFDLELLRYQN-VEEEMVQLLQLA 1013
Query: 1053 FTCLTESSQSRPTMEHVAKEL 1073
C RP++ V + +
Sbjct: 1014 VDCAAPYPDRRPSIAEVTRSI 1076
>BP032158
Length = 555
Score = 128 bits (322), Expect = 4e-30
Identities = 74/185 (40%), Positives = 102/185 (55%), Gaps = 2/185 (1%)
Frame = +2
Query: 791 GKMVYENILEATQDFDDKYLIGVGGQGSVYKAELHTGQVVAVKKLHPVSNEENLSPKSFT 850
G I AT +FD IG GG G VYK L G V+AVK+L S + N + F
Sbjct: 8 GYFSLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGDVIAVKQLSSKSKQGN---REFI 178
Query: 851 NEIQALTEIRHRNIVNLYGFCSHSQLSFLVYEFVEKGSLEKIL-KDDEEAIAFNWKKRVN 909
NEI ++ ++H N+V LYG C LVYE++E SL + L ++E+ + NW+ R+
Sbjct: 179 NEIGMISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMK 358
Query: 910 VIKDVANALCYMHHDCSPPIVHRDISSKNILLDSECVAHVSDFGTAKL-LDPNLTSSTSF 968
+ +A L Y+H + IVHRDI + N+LLD + A +SDFG AKL + N ST
Sbjct: 359 ICVGIAKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDEEENTHISTRI 538
Query: 969 ACTFG 973
A T G
Sbjct: 539 AGTIG 553
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 128 bits (322), Expect = 4e-30
Identities = 72/188 (38%), Positives = 109/188 (57%), Gaps = 2/188 (1%)
Frame = +1
Query: 792 KMVYENILEATQDFDDKYLIGVGGQGSVYKAELHTGQVVAVKKLHPVSNEENLSPKSFTN 851
K Y++I +ATQ+F ++G G G+VYKA + TG+VVAVK L P S + F
Sbjct: 196 KYSYKDIQKATQNFTT--ILGQGSFGTVYKATMPTGEVVAVKVLAPNSKQ---GEHEFQT 360
Query: 852 EIQALTEIRHRNIVNLYGFCSHSQLSFLVYEFVEKGSLEKILKDDEEAIAFNWKKRVNVI 911
E+ L + HRN+VNL GFC LVY+F+ GSL +L +E+ ++ W +R+ +
Sbjct: 361 EVHLLGRLHHRNLVNLVGFCVDKGQRILVYQFMSNGSLANLLYGEEKELS--WDERLQIA 534
Query: 912 KDVANALCYMHHDCSPPIVHRDISSKNILLDSECVAHVSDFGTAK--LLDPNLTSSTSFA 969
D+++ + Y+H PP++HRD+ S NILLD A V+DFG +K + D ++
Sbjct: 535 MDISHGIEYLHEGAVPPVIHRDLKSANILLDDSMRAMVADFGLSKEEIFDGR---NSGLK 705
Query: 970 CTFGYAAP 977
T+GY P
Sbjct: 706 GTYGYMDP 729
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,441,390
Number of Sequences: 28460
Number of extensions: 283213
Number of successful extensions: 2982
Number of sequences better than 10.0: 365
Number of HSP's better than 10.0 without gapping: 1885
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2475
length of query: 1083
length of database: 4,897,600
effective HSP length: 100
effective length of query: 983
effective length of database: 2,051,600
effective search space: 2016722800
effective search space used: 2016722800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC139747.2


