

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139744.6 + phase: 1 /partial
(384 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
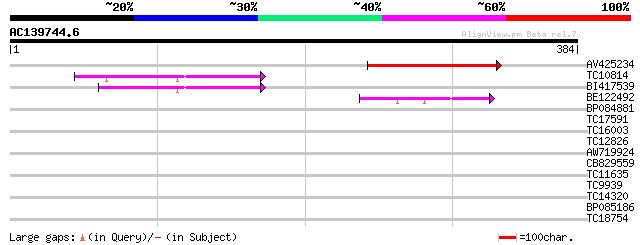
Score E
Sequences producing significant alignments: (bits) Value
AV425234 132 1e-31
TC10814 similar to UP|O22523 (O22523) DNA-binding protein GBP16,... 62 1e-10
BI417539 62 2e-10
BE122492 42 1e-04
BP084881 30 0.52
TC17591 similar to UP|Q9FLG1 (Q9FLG1) Beta-xylosidase, partial (... 28 2.0
TC16003 similar to UP|WR17_ARATH (Q9SJA8) Probable WRKY transcri... 27 4.4
TC12826 weakly similar to UP|Q9LVG7 (Q9LVG7) Ankyrin-like protei... 27 4.4
AW719924 27 4.4
CB829559 27 5.7
TC11635 similar to PIR|H86398|H86398 protein F17L21.10 [imported... 27 5.7
TC9939 homologue to UP|GSA_SOYBN (P45621) Glutamate-1-semialdehy... 27 5.7
TC14320 similar to UP|Q41111 (Q41111) Dehydrin, partial (56%) 27 7.5
BP085186 27 7.5
TC18754 similar to UP|Q9FZC3 (Q9FZC3) T1K7.25 protein, partial (... 26 9.7
>AV425234
Length = 280
Score = 132 bits (332), Expect = 1e-31
Identities = 62/91 (68%), Positives = 76/91 (83%)
Frame = +1
Query: 243 YRIHAGKTVPIVKGGEATVMEENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMPLR 302
Y+IHAGK+VPIVKGGE T MEE E++AIETF STG+G V +D++CSHYMKNFD G++PLR
Sbjct: 7 YQIHAGKSVPIVKGGEQTKMEEGEFFAIETFASTGKGYVREDLECSHYMKNFDVGHIPLR 186
Query: 303 LQSSKSLLSVINKNFSTLAFCKRWLDRAGCT 333
L +K LL+ INKNFSTLAF K++L R G T
Sbjct: 187 LPRAKQLLATINKNFSTLAFGKQYLYRLGET 279
>TC10814 similar to UP|O22523 (O22523) DNA-binding protein GBP16, partial
(31%)
Length = 525
Score = 62.4 bits (150), Expect = 1e-10
Identities = 42/139 (30%), Positives = 71/139 (50%), Gaps = 10/139 (7%)
Frame = +1
Query: 45 PTVNDSRTAKDRFTNEEKRA---LDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMI 101
PT+ +S ++++R LD + ++ + + AAE + + + KP ++
Sbjct: 106 PTLQESEVTGGATMSDDEREEKELDLTSPEVVTKYKSAAEIVNKALQLVISECKPKAKVV 285
Query: 102 QICEELENTAR-------KLIKEDGLKAGLAFPTGCSRNHCAAHYTPNAGDPTVLEYDDV 154
ICE+ ++ R K +K+ ++ G+AFPT S N+ H++P A D TVLE D+
Sbjct: 286 DICEKGDSYIREQTGSMYKNVKKK-IERGVAFPTCLSINNTVCHFSPLASDETVLEEGDL 462
Query: 155 TKIDFGTHINGRIIDCAFT 173
KID HI+G I A T
Sbjct: 463 LKIDMACHIDGFIAAVAHT 519
>BI417539
Length = 577
Score = 62.0 bits (149), Expect = 2e-10
Identities = 39/120 (32%), Positives = 64/120 (52%), Gaps = 7/120 (5%)
Frame = +2
Query: 61 EKRALDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTAR-------K 113
E++ LD + ++ + + AAE + + + KP ++ ICE+ ++ R K
Sbjct: 152 EEKELDLTSPEVVTKYKSAAEIVNKALQLVISECKPKAKVVDICEKGDSYIREQTGNMYK 331
Query: 114 LIKEDGLKAGLAFPTGCSRNHCAAHYTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFT 173
+K+ ++ G+AFPT S N+ H++P A D TVLE D+ KID HI+G I A T
Sbjct: 332 NVKKK-IERGVAFPTCLSINNTVCHFSPLASDETVLEEGDLLKIDMACHIDGFIAAVAHT 508
>BE122492
Length = 376
Score = 42.4 bits (98), Expect = 1e-04
Identities = 29/97 (29%), Positives = 47/97 (47%), Gaps = 6/97 (6%)
Frame = +3
Query: 238 HSISPYRIHAGKTVPIVKGGEATV----MEENEYYAIETFGSTGRG--QVHDDMDCSHYM 291
H + + I K V + V EENE YAI+ STG G ++ D+ + Y
Sbjct: 15 HQMKQFVIDGNKVVLSASNPDTRVDDAEFEENEVYAIDIVASTGEGKPKLLDEKQTTIYK 194
Query: 292 KNFDAGYMPLRLQSSKSLLSVINKNFSTLAFCKRWLD 328
+ D Y L++++S+ + S IN+ F + F R L+
Sbjct: 195 RAVDKSY-HLKMKASRFIFSEINQKFPIMPFSARALE 302
>BP084881
Length = 488
Score = 30.4 bits (67), Expect = 0.52
Identities = 20/55 (36%), Positives = 28/55 (50%), Gaps = 6/55 (10%)
Frame = -3
Query: 194 GIKAAGIDVP----LCEIGAAIQEVMESYEVELDGKTYQVKSIRNL--NGHSISP 242
G+ + G+ P +C I QEVM V DG TY+ ++IR +GH SP
Sbjct: 456 GLSSEGLHQPPSYFICPI---FQEVMRDPHVAADGFTYEAEAIRGWLDSGHDTSP 301
>TC17591 similar to UP|Q9FLG1 (Q9FLG1) Beta-xylosidase, partial (24%)
Length = 582
Score = 28.5 bits (62), Expect = 2.0
Identities = 12/37 (32%), Positives = 20/37 (53%)
Frame = +3
Query: 284 DMDCSHYMKNFDAGYMPLRLQSSKSLLSVINKNFSTL 320
D+DC Y+ + G + L S+ + I+ NF+TL
Sbjct: 144 DLDCGSYLGKYTEGAVKQGLLDEASINNAISNNFATL 254
>TC16003 similar to UP|WR17_ARATH (Q9SJA8) Probable WRKY transcription
factor 17 (WRKY DNA-binding protein 17), partial (20%)
Length = 446
Score = 27.3 bits (59), Expect = 4.4
Identities = 12/17 (70%), Positives = 13/17 (75%)
Frame = -1
Query: 1 GGEAQKKKKNKKKKGKK 17
GG +KKKK KKKK KK
Sbjct: 182 GGYGRKKKKKKKKKKKK 132
>TC12826 weakly similar to UP|Q9LVG7 (Q9LVG7) Ankyrin-like protein, partial
(23%)
Length = 528
Score = 27.3 bits (59), Expect = 4.4
Identities = 11/34 (32%), Positives = 18/34 (52%)
Frame = +1
Query: 129 GCSRNHCAAHYTPNAGDPTVLEYDDVTKIDFGTH 162
GC R HC A+ P + + +Y +VT++ H
Sbjct: 133 GCQRKHCTAYSNP---EESSKDYSEVTRLQ*NRH 225
>AW719924
Length = 505
Score = 27.3 bits (59), Expect = 4.4
Identities = 12/18 (66%), Positives = 14/18 (77%)
Frame = -1
Query: 3 EAQKKKKNKKKKGKKVQT 20
E +KKKK KKKK KK +T
Sbjct: 55 EKKKKKKKKKKKKKKART 2
>CB829559
Length = 552
Score = 26.9 bits (58), Expect = 5.7
Identities = 17/61 (27%), Positives = 24/61 (38%)
Frame = +2
Query: 81 EAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGLAFPTGCSRNHCAAHYT 140
EA T + W+ ++Q+CE + R L+ TGC NH A Y
Sbjct: 245 EASHPTGTMLSHWVP--CNLLQVCEMFRKSLRWLL------------TGCILNHSLA*YL 382
Query: 141 P 141
P
Sbjct: 383 P 385
>TC11635 similar to PIR|H86398|H86398 protein F17L21.10 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(98%)
Length = 633
Score = 26.9 bits (58), Expect = 5.7
Identities = 10/20 (50%), Positives = 15/20 (75%)
Frame = +3
Query: 6 KKKKNKKKKGKKVQTDPPTI 25
KKKK KKKK ++++ DP +
Sbjct: 3 KKKKKKKKKKREIKMDPDAL 62
>TC9939 homologue to UP|GSA_SOYBN (P45621) Glutamate-1-semialdehyde
2,1-aminomutase, chloroplast precursor (GSA)
(Glutamate-1-semialdehyde aminotransferase) (GSA-AT) ,
partial (52%)
Length = 903
Score = 26.9 bits (58), Expect = 5.7
Identities = 14/41 (34%), Positives = 17/41 (41%)
Frame = +1
Query: 126 FPTGCSRNHCAAHYTPNAGDPTVLEYDDVTKIDFGTHINGR 166
FP+ SRN C H +PT + T H NGR
Sbjct: 61 FPSQSSRNGCFRHQIHTHRNPTSPPFSSTTTNSNSFHHNGR 183
>TC14320 similar to UP|Q41111 (Q41111) Dehydrin, partial (56%)
Length = 1231
Score = 26.6 bits (57), Expect = 7.5
Identities = 11/26 (42%), Positives = 17/26 (65%)
Frame = +3
Query: 2 GEAQKKKKNKKKKGKKVQTDPPTIPI 27
GE +KKKK K+KK K + ++P+
Sbjct: 639 GEEKKKKKKKEKKEVKSTDENTSVPV 716
>BP085186
Length = 388
Score = 26.6 bits (57), Expect = 7.5
Identities = 12/17 (70%), Positives = 13/17 (75%)
Frame = -2
Query: 1 GGEAQKKKKNKKKKGKK 17
G E +KKKK KKKK KK
Sbjct: 270 GKEMKKKKKKKKKKKKK 220
>TC18754 similar to UP|Q9FZC3 (Q9FZC3) T1K7.25 protein, partial (37%)
Length = 873
Score = 26.2 bits (56), Expect = 9.7
Identities = 16/57 (28%), Positives = 27/57 (47%)
Frame = +1
Query: 76 VRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGLAFPTGCSR 132
+R A + HRQ + H+QQ G+ + +++ R L+ L A + P G R
Sbjct: 529 LRHACDIHRQIKGHVQQM---GLNLASCGDDMLQFRRCLVASFFLNAAIKQPDGTYR 690
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,466,346
Number of Sequences: 28460
Number of extensions: 83766
Number of successful extensions: 694
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 515
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 615
length of query: 384
length of database: 4,897,600
effective HSP length: 92
effective length of query: 292
effective length of database: 2,279,280
effective search space: 665549760
effective search space used: 665549760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC139744.6


