

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139708.6 - phase: 0
(272 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
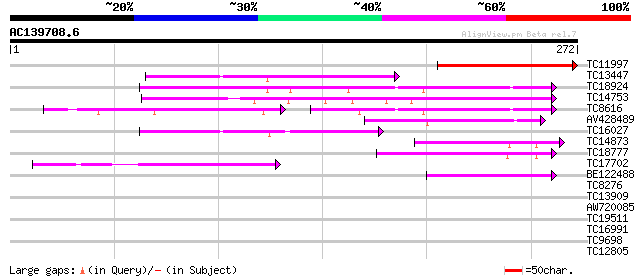
Score E
Sequences producing significant alignments: (bits) Value
TC11997 similar to UP|O48940 (O48940) Polyphosphoinositide bindi... 128 9e-31
TC13447 82 1e-16
TC18924 homologue to UP|Q94FN3 (Q94FN3) Phosphatidylinositol tra... 81 2e-16
TC14753 similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial... 80 5e-16
TC8616 UP|Q94FN1 (Q94FN1) Phosphatidylinositol transfer-like pro... 52 4e-15
AV428489 52 1e-07
TC16027 47 3e-06
TC14873 weakly similar to GB|BAB08982.1|9758453|AB008271 seleniu... 46 8e-06
TC18777 weakly similar to UP|Q9LDJ7 (Q9LDJ7) ESTs AU081555, part... 44 4e-05
TC17702 weakly similar to GB|AAN73306.1|25141223|BT002309 At3g51... 41 2e-04
BE122488 40 3e-04
TC8276 similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial ... 34 0.030
TC13909 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, ... 32 0.15
AW720085 31 0.26
TC19511 similar to UP|Q9ARI4 (Q9ARI4) PII protein, partial (66%) 31 0.26
TC16991 similar to GB|AAN73306.1|25141223|BT002309 At3g51670/T18... 27 3.7
TC9698 similar to UP|AAR83120 (AAR83120) Chloroplast carotenoid ... 27 4.8
TC12805 26 8.3
>TC11997 similar to UP|O48940 (O48940) Polyphosphoinositide binding protein
Ssh2p, partial (26%)
Length = 492
Score = 128 bits (322), Expect = 9e-31
Identities = 59/67 (88%), Positives = 67/67 (99%)
Frame = +2
Query: 206 KLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGGKLP 265
KLFIVHAPY+FMKVWK++YPFIDD+TKKKIVFV+N+KLK+TLLEEIDESQLPEIYGG+LP
Sbjct: 2 KLFIVHAPYIFMKVWKLVYPFIDDHTKKKIVFVDNQKLKSTLLEEIDESQLPEIYGGQLP 181
Query: 266 LVPIQDS 272
LVPIQDS
Sbjct: 182 LVPIQDS 202
>TC13447
Length = 554
Score = 81.6 bits (200), Expect = 1e-16
Identities = 44/123 (35%), Positives = 71/123 (56%), Gaps = 1/123 (0%)
Frame = +2
Query: 66 DDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQG-LD 124
D+ MI+ FL+ R VD+A + K +KWR+ F S ++ + D K YV LD
Sbjct: 176 DEDMIQWFLKDRKFSVDEAVSKLTKAIKWRQDFEVS-KLTEEAVKDAARSGKAYVHDFLD 352
Query: 125 KKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPGEEKFVSIADIKGWGYANS 184
GRP++V A+KHF +R VF +EK +S++P G+E+ + I D++G+G N+
Sbjct: 353 INGRPVLVVVASKHFPKAQDPSDDERLCVFLVEKALSKLPSGKEQVLGIVDLRGFGTENA 532
Query: 185 DIR 187
D++
Sbjct: 533 DLK 541
>TC18924 homologue to UP|Q94FN3 (Q94FN3) Phosphatidylinositol transfer-like
protein II, complete
Length = 1976
Score = 81.3 bits (199), Expect = 2e-16
Identities = 65/219 (29%), Positives = 99/219 (44%), Gaps = 19/219 (8%)
Frame = +3
Query: 63 KEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQG 122
K D + RFL+AR D +K+ M+ ++WRK F V + + K Y G
Sbjct: 264 KHDDYHQLLRFLKARKFDTEKSKQMWSDMLQWRKEFGADTIVEDFDFKEIDEVVKYYPHG 443
Query: 123 ---LDKKGRPIIVA----FAAKHFQNKNGLDAFKRYVVFALEKLIS--------RMPPGE 167
+DK GRP+ + A +D + +Y V E+
Sbjct: 444 HHGVDKDGRPVYIENIGQVDATKLMQVTTMDRYIKYHVKEFERTFDLKFAACSIAAKKHI 623
Query: 168 EKFVSIADIKGWGYANSDIRGYLGALTILQ----DYYPERLGKLFIVHAPYMFMKVWKII 223
++ +I D++G G N + + +T LQ D YPE L ++FI++A F +W +
Sbjct: 624 DQSTTILDVQGVGLKNFN-KHARELITRLQKIDGDNYPETLNRMFIINAGSGFRMLWSTV 800
Query: 224 YPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
F+D T KI + N K ++ LLE ID SQLPE GG
Sbjct: 801 KSFLDPKTTSKIHVLGN-KYQSKLLEVIDASQLPEFLGG 914
>TC14753 similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial (57%)
Length = 1472
Score = 79.7 bits (195), Expect = 5e-16
Identities = 65/218 (29%), Positives = 96/218 (43%), Gaps = 19/218 (8%)
Frame = +1
Query: 64 EVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQE---KIYV 120
E D+++ +FLRARD V A M ++WRK F V +DL E ++
Sbjct: 73 ERSDVVLLKFLRARDFKVKDAFTMIKNTVRWRKEFGIDALVD-----EDLGSEWDKVVFT 237
Query: 121 QGLDKKGRPIIV----AFAAKHFQNKNGLDAFKR-----YVVFALEKLISRM-----PPG 166
QG DK G + F K K D KR + + LEK + ++ P
Sbjct: 238 QGHDKLGHAVCYNVFGEFEDKELYQKTFSDDEKRQKFIKWRIQVLEKSVRKLDFSPTPTA 417
Query: 167 EEKFVSIADIKGW-GYANSDIRGYLG-ALTILQDYYPERLGKLFIVHAPYMFMKVWKIIY 224
V + D+K G ++R AL +LQD YPE + K ++ P+ ++ ++I
Sbjct: 418 ISTIVQVNDLKNSPGLGKWELRQATNQALQLLQDNYPEFVAKQVFINVPWWYLAFSRMIS 597
Query: 225 PFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
PF+ TK K VF K TL + I +P YGG
Sbjct: 598 PFLTQRTKSKFVFAGPSKSAETLFKYIAPELVPVQYGG 711
>TC8616 UP|Q94FN1 (Q94FN1) Phosphatidylinositol transfer-like protein III,
complete
Length = 2302
Score = 51.6 bits (122), Expect(2) = 4e-15
Identities = 42/130 (32%), Positives = 61/130 (46%), Gaps = 12/130 (9%)
Frame = +3
Query: 145 LDAFKRYVVFALEKLISRMPPG--------EEKFVSIADIKGWGYANSDIRGYLGALTIL 196
+D + RY V EK + P + +I D++G G N + +T L
Sbjct: 654 MDRYVRYHVQEFEKSFAIKFPACTMAAKRHIDSSTTILDVQGVGLKNFT-KSARELITRL 830
Query: 197 Q----DYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATLLEEID 252
Q D YPE L ++FI++A F +W + F+D T KI + N K + LE ID
Sbjct: 831 QKVDGDNYPETLCQMFIINAGPGFRLLWNTVKSFLDPKTTSKIHVLGN-KYHSKWLEVID 1007
Query: 253 ESQLPEIYGG 262
S+LPE GG
Sbjct: 1008ASELPECLGG 1037
Score = 45.1 bits (105), Expect(2) = 4e-15
Identities = 34/126 (26%), Positives = 60/126 (46%), Gaps = 10/126 (7%)
Frame = +2
Query: 17 KHELAKEETKGKILVQDDGALNDST------EAELTKINLMRTLVESRDPSSKEVDDL-M 69
KH L K+ ++ K DG ++ + EL ++ R + + + DD M
Sbjct: 239 KHSLRKKSSRRK----SDGRVSSVSIEDVRDVEELQAVDAFRQSLIMDELLPQAFDDYHM 406
Query: 70 IRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYV---QGLDKK 126
+ RFL+AR D++KA M+ + ++WRK F + E + + Y G+DK+
Sbjct: 407 MLRFLKARKFDIEKAKHMWAEMLQWRKEFGADTIMQDFEFQELDEVVRYYPHGHHGVDKE 586
Query: 127 GRPIIV 132
GRP+ +
Sbjct: 587 GRPVYI 604
>AV428489
Length = 386
Score = 51.6 bits (122), Expect = 1e-07
Identities = 27/88 (30%), Positives = 50/88 (56%), Gaps = 1/88 (1%)
Frame = +1
Query: 171 VSIADIKGWGYANSDIRGYLGALTILQDY-YPERLGKLFIVHAPYMFMKVWKIIYPFIDD 229
V + D+ G ++ + L A++ + D YPE+ +IV+APY+F WK++ P + +
Sbjct: 16 VKVLDMSGLKFSALNQLRLLTAISTVDDLNYPEKTDTYYIVNAPYVFSACWKVVKPLLQE 195
Query: 230 NTKKKIVFVENKKLKATLLEEIDESQLP 257
T++KI ++ K LL+ +D + LP
Sbjct: 196 RTRRKIQVLQGSG-KEELLKIMDYASLP 276
>TC16027
Length = 653
Score = 47.4 bits (111), Expect = 3e-06
Identities = 31/118 (26%), Positives = 59/118 (49%), Gaps = 1/118 (0%)
Frame = +2
Query: 63 KEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQG 122
K D +RR+L AR+ +VDK+ M + +KWR ++ P + +E+A + K+
Sbjct: 305 KYCTDACLRRYLEARNWNVDKSKKMLEETLKWRSTYKPE-EIRWAEVAHEGETGKVSRAN 481
Query: 123 L-DKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPGEEKFVSIADIKGW 179
D+ GR +++ QN + +++V+ LE I + G+E+ + D G+
Sbjct: 482 FHDRLGRTVLI--LRPGMQNTASPEDNIKHLVYLLENAILNLSEGQEQMSWLIDFTGF 649
>TC14873 weakly similar to GB|BAB08982.1|9758453|AB008271 selenium-binding
protein-like {Arabidopsis thaliana;} , partial (7%)
Length = 763
Score = 45.8 bits (107), Expect = 8e-06
Identities = 27/74 (36%), Positives = 40/74 (53%), Gaps = 2/74 (2%)
Frame = +2
Query: 195 ILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFV-ENKKLKATLLEEI-D 252
IL +YPERL F+ + P +F +K + F+D T +K+ FV N K L++ + D
Sbjct: 68 ILLYHYPERLAIAFLFNPPSIFQAFYKAVKYFLDPKTAQKVKFVYPNNKDSVELMKSLFD 247
Query: 253 ESQLPEIYGGKLPL 266
LP +GGK L
Sbjct: 248 IDNLPSEFGGKATL 289
>TC18777 weakly similar to UP|Q9LDJ7 (Q9LDJ7) ESTs AU081555, partial (24%)
Length = 693
Score = 43.5 bits (101), Expect = 4e-05
Identities = 24/88 (27%), Positives = 44/88 (49%), Gaps = 2/88 (2%)
Frame = +1
Query: 177 KGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIV 236
+G+ +N + + +LQ+YYPERLG I +AP++F ++ P + K+
Sbjct: 1 QGFNLSNISFKLARETIHVLQEYYPERLGLSLIYNAPWIFQPFLAMVKPLLQPENYNKLK 180
Query: 237 F-VENKKLKATLLEEI-DESQLPEIYGG 262
F N + ++E++ D L +GG
Sbjct: 181 FGYSNDEGTKKIMEDLFDMDHLESAFGG 264
>TC17702 weakly similar to GB|AAN73306.1|25141223|BT002309
At3g51670/T18N14_50 {Arabidopsis thaliana;}, partial
(20%)
Length = 574
Score = 40.8 bits (94), Expect = 2e-04
Identities = 33/119 (27%), Positives = 53/119 (43%)
Frame = +2
Query: 12 SALDSKHELAKEETKGKILVQDDGALNDSTEAELTKINLMRTLVESRDPSSKEVDDLMIR 71
S L + + A +E K K+L G +N+++ +T +N + D+++
Sbjct: 245 SHLRNSEKKALQEFKDKLLAS--GEVNNASMWGVTLLN------------GDDRADVLLL 382
Query: 72 RFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGLDKKGRPI 130
+FLRARD V A M LK + WRK F V +L Q D++G P+
Sbjct: 383 KFLRARDFRVCDALNMLLKCLAWRKEFGADNIVDEELGFKELEGVVACTQVYDREGHPV 559
>BE122488
Length = 346
Score = 40.4 bits (93), Expect = 3e-04
Identities = 19/62 (30%), Positives = 32/62 (50%)
Frame = +2
Query: 201 PERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIY 260
PE + K ++ P+ + + +I PF+ TK K V K+ TL++ I ++P Y
Sbjct: 17 PELVAKNIFINVPFWYYALNALISPFLTQRTKSKFVIGRPAKVTETLIKYIPIEEIPVHY 196
Query: 261 GG 262
GG
Sbjct: 197 GG 202
>TC8276 similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial (42%)
Length = 1071
Score = 33.9 bits (76), Expect = 0.030
Identities = 17/43 (39%), Positives = 22/43 (50%)
Frame = +3
Query: 220 WKIIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
+ +I PF+ TK K VF K TL + I Q+P YGG
Sbjct: 21 YTMINPFLTQRTKSKFVFAGPSKSPDTLFKYISPEQVPIQYGG 149
>TC13909 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial
(20%)
Length = 399
Score = 31.6 bits (70), Expect = 0.15
Identities = 22/92 (23%), Positives = 41/92 (43%), Gaps = 8/92 (8%)
Frame = +3
Query: 15 DSKHELAKEETKGKILVQDDGALNDSTEAELTKINLMRTLVESRDPSS--------KEVD 66
+ H+L + G Q+ D +++E + ++ L+ D S +
Sbjct: 120 EEDHQLKIPQNLGSFK-QESNLPCDLSDSETKAVQDLKQLLTVTDEPSIWGVPLLKDDRT 296
Query: 67 DLMIRRFLRARDLDVDKASAMFLKYMKWRKSF 98
D+++ +FLRAR+ V A M ++WRK F
Sbjct: 297 DVILLKFLRAREFKVKDALLMINNTLRWRKDF 392
>AW720085
Length = 524
Score = 30.8 bits (68), Expect = 0.26
Identities = 18/66 (27%), Positives = 32/66 (48%), Gaps = 5/66 (7%)
Frame = +1
Query: 70 IRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQ-----EKIYVQGLD 124
+ RFL+AR+ + KA M + + WR +S I +DL + + I + G
Sbjct: 235 LMRFLKAREWNASKAHKMLVDCLNWRVQSEIDSILSKPIIPEDLYRAVRDSQLIGLSGFS 414
Query: 125 KKGRPI 130
++G P+
Sbjct: 415 REGLPV 432
>TC19511 similar to UP|Q9ARI4 (Q9ARI4) PII protein, partial (66%)
Length = 631
Score = 30.8 bits (68), Expect = 0.26
Identities = 16/49 (32%), Positives = 30/49 (60%), Gaps = 3/49 (6%)
Frame = +1
Query: 226 FIDDNTKKKI---VFVENKKLKATLLEEIDESQLPEIYGGKLPLVPIQD 271
F +DN K+ + V N +++A + + I+E++ EI GK+ L+P+ D
Sbjct: 349 FSEDNFVAKVKMEIVVRNDQVEAVIDKIIEEARTGEIGDGKIFLIPVSD 495
>TC16991 similar to GB|AAN73306.1|25141223|BT002309 At3g51670/T18N14_50
{Arabidopsis thaliana;}, partial (38%)
Length = 681
Score = 26.9 bits (58), Expect = 3.7
Identities = 12/38 (31%), Positives = 18/38 (46%)
Frame = +3
Query: 225 PFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
PF+ TK K V + TL + + ++P YGG
Sbjct: 6 PFLTQRTKSKFVISKEGNAAETLYKFVRPEEIPVRYGG 119
>TC9698 similar to UP|AAR83120 (AAR83120) Chloroplast carotenoid
epsilon-ring hydroxylase, partial (19%)
Length = 620
Score = 26.6 bits (57), Expect = 4.8
Identities = 14/28 (50%), Positives = 18/28 (64%)
Frame = -1
Query: 78 DLDVDKASAMFLKYMKWRKSFVPSGSVS 105
D D+DK +A LK M W+ + P GSVS
Sbjct: 380 D*DIDKFTAS-LKKMNWKLTISPVGSVS 300
>TC12805
Length = 702
Score = 25.8 bits (55), Expect = 8.3
Identities = 14/33 (42%), Positives = 18/33 (54%)
Frame = +1
Query: 149 KRYVVFALEKLISRMPPGEEKFVSIADIKGWGY 181
+RY V + KL RMP G + S+ D G GY
Sbjct: 178 ERYAVTDMRKLAHRMPFGIPEESSLGDGLGEGY 276
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.137 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,754,905
Number of Sequences: 28460
Number of extensions: 40432
Number of successful extensions: 192
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 188
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 188
length of query: 272
length of database: 4,897,600
effective HSP length: 89
effective length of query: 183
effective length of database: 2,364,660
effective search space: 432732780
effective search space used: 432732780
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 54 (25.4 bits)
Medicago: description of AC139708.6


