

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139708.14 - phase: 0
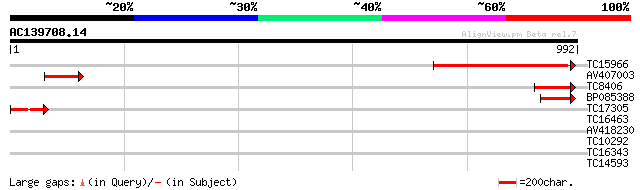
(992 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC15966 similar to UP|Q8L5C2 (Q8L5C2) 110 kDa 4SNc-tudor protein... 421 e-118
AV407003 125 4e-29
TC8406 similar to UP|Q8L5C2 (Q8L5C2) 110 kDa 4SNc-tudor protein,... 121 5e-28
BP085388 103 2e-22
TC17305 weakly similar to UP|Q9FLT0 (Q9FLT0) Transcription facto... 100 1e-21
TC16463 similar to UP|AAR87242 (AAR87242) Expressed protein, par... 30 1.9
AV418230 30 1.9
TC10292 homologue to UP|Q8GSM8 (Q8GSM8) Squalene monooxygenase 1... 30 2.5
TC16343 similar to UP|Q8LB75 (Q8LB75) Galactose-1-phosphate urid... 29 3.2
TC14593 similar to UP|Q8H1U3 (Q8H1U3) CDC26, partial (31%) 28 9.3
>TC15966 similar to UP|Q8L5C2 (Q8L5C2) 110 kDa 4SNc-tudor protein, partial
(25%)
Length = 891
Score = 421 bits (1083), Expect = e-118
Identities = 211/249 (84%), Positives = 224/249 (89%)
Frame = +1
Query: 742 EVLGGGKFYVQTVGDQKIASIQNQLASLNLKDAPVIGAFNPKKGDIVLCYFHADSSWYRA 801
EVLGG KFYVQTVGDQKIASIQ QLASLNLK+APV+GAF+PKKGD VLCYFH D SWYRA
Sbjct: 1 EVLGGDKFYVQTVGDQKIASIQQQLASLNLKEAPVLGAFSPKKGDTVLCYFHGDKSWYRA 180
Query: 802 MVVNTPRGPVESSKDAFEVFYIDYGNQEVVPYSQLRPLDPSVSAAPGLAQLCSLAYIKLP 861
MVVNTPRGPVES +D FEVFYIDYGNQE V YSQLRPLD SVSAAPGLAQLCSLAYIK P
Sbjct: 181 MVVNTPRGPVESPQDIFEVFYIDYGNQEQVAYSQLRPLDQSVSAAPGLAQLCSLAYIKSP 360
Query: 862 NLEEDFGQEAAEYLSELTLSSGKEFRAMVEEKDTTGGKVKGQGTGPIIAVTLVAVDSEIS 921
+LEEDFGQEAAEYLSELTLSSGKEFRA VEE+DT+GGK KGQGTG I+AVTLVAVD+EIS
Sbjct: 361 SLEEDFGQEAAEYLSELTLSSGKEFRAQVEERDTSGGKAKGQGTGTILAVTLVAVDAEIS 540
Query: 922 VNAAMLQEGLARMEKRNRWDRTARKQALDNLEMFQGEARTARRGMWQYGDIQSDDEDTAP 981
VNAAMLQEGLARMEKRNRWDR RK LD+LE FQ EART RRGMWQYGD++SD+ED P
Sbjct: 541 VNAAMLQEGLARMEKRNRWDRKERKAGLDSLEKFQDEARTKRRGMWQYGDVESDEED-GP 717
Query: 982 PQRKAGGGR 990
P RKAG GR
Sbjct: 718 PARKAGTGR 744
>AV407003
Length = 222
Score = 125 bits (313), Expect = 4e-29
Identities = 58/68 (85%), Positives = 64/68 (93%)
Frame = +1
Query: 62 EPFAWESREFLRKLLIGKEITFRIDYTVPSINREFGTVFLGDKNVALLVVSQGWAKVREQ 121
EPFAWESRE+LRKL IGKE+TFR+DY+V SINR+FGTVFLGDKNV +LVVSQGWAKVREQ
Sbjct: 19 EPFAWESREYLRKLCIGKEVTFRVDYSVASINRDFGTVFLGDKNVGVLVVSQGWAKVREQ 198
Query: 122 GQQKGEAS 129
GQQKGE S
Sbjct: 199 GQQKGEVS 222
Score = 30.4 bits (67), Expect = 1.4
Identities = 18/54 (33%), Positives = 29/54 (53%)
Frame = +1
Query: 434 PYAREAKEFLRTRLIGRQVNVQMEYSRKVGPVDGSAVPPGAVDSRVMDFGSVFV 487
P+A E++E+LR IG++V +++YS V S DFG+VF+
Sbjct: 22 PFAWESREYLRKLCIGKEVTFRVDYS---------------VASINRDFGTVFL 138
>TC8406 similar to UP|Q8L5C2 (Q8L5C2) 110 kDa 4SNc-tudor protein, partial
(7%)
Length = 562
Score = 121 bits (304), Expect = 5e-28
Identities = 57/72 (79%), Positives = 62/72 (85%)
Frame = +1
Query: 919 EISVNAAMLQEGLARMEKRNRWDRTARKQALDNLEMFQGEARTARRGMWQYGDIQSDDED 978
EISVNAAMLQEGLARMEKRNRWDR RK LD+L+ FQ +AR RRGMWQYGD++SDDED
Sbjct: 1 EISVNAAMLQEGLARMEKRNRWDRKERKVGLDSLQKFQDDARKERRGMWQYGDVESDDED 180
Query: 979 TAPPQRKAGGGR 990
TAPP RKAG GR
Sbjct: 181 TAPPARKAGAGR 216
>BP085388
Length = 465
Score = 103 bits (256), Expect = 2e-22
Identities = 47/62 (75%), Positives = 51/62 (81%)
Frame = -3
Query: 929 EGLARMEKRNRWDRTARKQALDNLEMFQGEARTARRGMWQYGDIQSDDEDTAPPQRKAGG 988
EGLARMEKRNRWDR RK LD+LE FQ +AR RRGMWQYGD++SDDED APP RKAG
Sbjct: 463 EGLARMEKRNRWDRKERKVGLDSLEKFQDDARKERRGMWQYGDVESDDEDDAPPARKAGA 284
Query: 989 GR 990
GR
Sbjct: 283 GR 278
>TC17305 weakly similar to UP|Q9FLT0 (Q9FLT0) Transcription factor-like
protein (100 kDa coactivator-like protein), partial (5%)
Length = 616
Score = 100 bits (249), Expect = 1e-21
Identities = 48/67 (71%), Positives = 58/67 (85%)
Frame = -3
Query: 1 MAATAAGNSAWYKAKVKAVPSGDCIVVVSVAANAKLGVLPEKSITLSSLIAPRLARRGGV 60
MA+ A G + WY+ +VKAVPSGDC+V+V+VA++ K G LPEKSITL+SLI PRLARRGGV
Sbjct: 200 MASAATGATGWYRGRVKAVPSGDCLVIVAVASS-KPGPLPEKSITLASLITPRLARRGGV 24
Query: 61 DEPFAWE 67
DEPFAWE
Sbjct: 23 DEPFAWE 3
Score = 32.0 bits (71), Expect = 0.49
Identities = 17/57 (29%), Positives = 30/57 (51%)
Frame = -3
Query: 382 FTGKVVEVVSGDCVIVADDSIPYGSPQAERRVNLSSIRCPKMGNPRRDEKPAPYARE 438
+ G+V V SGDC+++ + P E+ + L+S+ P++ RR P+A E
Sbjct: 167 YRGRVKAVPSGDCLVIVAVASSKPGPLPEKSITLASLITPRLA--RRGGVDEPFAWE 3
>TC16463 similar to UP|AAR87242 (AAR87242) Expressed protein, partial (92%)
Length = 640
Score = 30.0 bits (66), Expect = 1.9
Identities = 14/32 (43%), Positives = 19/32 (58%), Gaps = 1/32 (3%)
Frame = +3
Query: 659 TFLGSLWESRANGAVPLLEAGLA-KLQTSFGS 689
T++G LW R A P+L+ GL + SFGS
Sbjct: 93 TWIGHLWRPRLQAAKPILQRGLVYSISLSFGS 188
>AV418230
Length = 405
Score = 30.0 bits (66), Expect = 1.9
Identities = 20/47 (42%), Positives = 25/47 (52%)
Frame = +2
Query: 163 NLPPSALGDASNFDAMGLLAKNKGVPMEALVEQVRDGSTLRIYLLPE 209
NLPP LGD S+ A+ L+A G+P AL + DG I PE
Sbjct: 8 NLPPRPLGDRSHPTAVVLVASPGGLP-PALFCRWGDGGLAEIRAAPE 145
>TC10292 homologue to UP|Q8GSM8 (Q8GSM8) Squalene monooxygenase 1 , partial
(33%)
Length = 663
Score = 29.6 bits (65), Expect = 2.5
Identities = 21/60 (35%), Positives = 31/60 (51%)
Frame = -1
Query: 413 VNLSSIRCPKMGNPRRDEKPAPYAREAKEFLRTRLIGRQVNVQMEYSRKVGPVDGSAVPP 472
+NLS I R ++P PY AK++ RT+L GR N + S++ GP + PP
Sbjct: 402 INLSPIHMRLGEGNGRSKRPTPYIATAKKW-RTKLKGRGFNPE---SKETGPSENK--PP 241
>TC16343 similar to UP|Q8LB75 (Q8LB75) Galactose-1-phosphate uridyl
transferase-like protein, partial (28%)
Length = 466
Score = 29.3 bits (64), Expect = 3.2
Identities = 19/45 (42%), Positives = 23/45 (50%), Gaps = 9/45 (20%)
Frame = +2
Query: 249 PAEPRAPLTSAQR---------LAVSASAAETSADPFGADAKFFT 284
P EP APL S+Q LA SASA TSA P + ++ T
Sbjct: 116 PREPNAPLISSQNHPPTRTRTPLAPSASATSTSAHPRSSGSRPMT 250
>TC14593 similar to UP|Q8H1U3 (Q8H1U3) CDC26, partial (31%)
Length = 1112
Score = 27.7 bits (60), Expect = 9.3
Identities = 22/84 (26%), Positives = 38/84 (45%), Gaps = 10/84 (11%)
Frame = -2
Query: 897 GGKVKGQGTGPIIAVTLVAVDSEISVNAAMLQ------EGLARMEKRNR----WDRTARK 946
GG V+G G +AV+ ++E+ ++ + + +G ME+R + W R R+
Sbjct: 337 GGDVEGGEQGGDLAVSFGVGEAELDLDLHIGRGERKGADGEGFMERRGQCDALWKRDPRR 158
Query: 947 QALDNLEMFQGEARTARRGMWQYG 970
D E + RR W+YG
Sbjct: 157 DRTDE*EWRRQWWLLRRRWRWRYG 86
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,801,928
Number of Sequences: 28460
Number of extensions: 161977
Number of successful extensions: 766
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 757
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 763
length of query: 992
length of database: 4,897,600
effective HSP length: 99
effective length of query: 893
effective length of database: 2,080,060
effective search space: 1857493580
effective search space used: 1857493580
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC139708.14


