

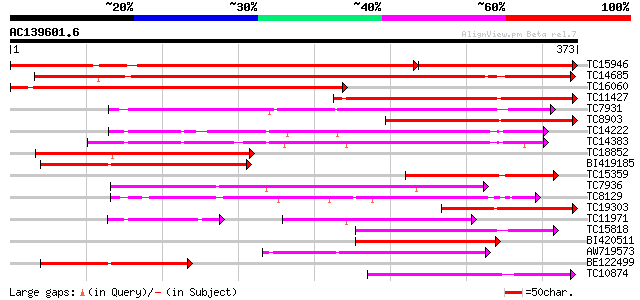
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139601.6 - phase: 0
(373 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC15946 weakly similar to UP|Q43383 (Q43383) 2A6 protein, partia... 390 e-152
TC14685 weakly similar to UP|Q43383 (Q43383) 2A6 protein, partia... 290 3e-79
TC16060 weakly similar to UP|Q9SHK4 (Q9SHK4) F12K11.6, partial (8%) 275 9e-75
TC11427 weakly similar to UP|Q9LTH7 (Q9LTH7) Leucoanthocyanidin ... 170 3e-43
TC7931 weakly similar to UP|Q43640 (Q43640) Dioxygenase, partial... 160 2e-40
TC8903 weakly similar to UP|Q9LSW7 (Q9LSW7) 1-aminocyclopropane-... 159 7e-40
TC14222 similar to UP|Q41681 (Q41681) 1-aminocyclopropane-1-carb... 156 5e-39
TC14383 homologue to UP|Q8W3Y3 (Q8W3Y3) 1-aminocyclopropane-1-ca... 149 1e-36
TC18852 weakly similar to UP|Q9SHK4 (Q9SHK4) F12K11.6, partial (4%) 146 6e-36
BI419185 132 9e-32
TC15359 weakly similar to UP|Q84MB3 (Q84MB3) At1g06620, partial ... 131 2e-31
TC7936 similar to UP|Q9SP53 (Q9SP53) Flavanone 3-hydroxylase, pa... 130 3e-31
TC8129 similar to UP|Q9XG83 (Q9XG83) GA 2-oxidase, partial (90%) 116 7e-27
TC19303 similar to UP|Q84MB3 (Q84MB3) At1g06620, partial (15%) 106 7e-24
TC11971 weakly similar to UP|Q43419 (Q43419) ACC oxidase, partia... 87 1e-22
TC15818 similar to UP|Q9FFF6 (Q9FFF6) Leucoanthocyanidin dioxyge... 100 3e-22
BI420511 100 5e-22
AW719573 99 1e-21
BE122499 97 4e-21
TC10874 similar to UP|Q8LF06 (Q8LF06) Leucoanthocyanidin dioxyge... 91 4e-19
>TC15946 weakly similar to UP|Q43383 (Q43383) 2A6 protein, partial (56%)
Length = 1252
Score = 390 bits (1002), Expect(2) = e-152
Identities = 193/272 (70%), Positives = 228/272 (82%), Gaps = 3/272 (1%)
Frame = +1
Query: 1 MVVENSNQLKESSDFT-YDREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRK-LEISES 58
MVV+N+N+L+E +D T YDREAEVKAFDD+K GVKGLV+ GV+KIPRMF+ K L+I
Sbjct: 31 MVVKNTNKLEEVTDSTHYDREAEVKAFDDAKTGVKGLVECGVTKIPRMFHANKHLDI--- 201
Query: 59 TASDSKLS-IPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDG 117
T +SKLS +P++DL D H EVI +IRSACHEWGFFQVINH IP VLDEMIDG
Sbjct: 202 TPENSKLSSVPLIDLTDEHS------EVIGKIRSACHEWGFFQVINHGIPTSVLDEMIDG 363
Query: 118 ICRFHEQDADVRKEFYTRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEVPQ 177
I RFHEQD++VRK+F++RDLKKKV YY+N+ LF+GQ ANWRDTFGFA+APDP PDE+P
Sbjct: 364 IRRFHEQDSEVRKQFHSRDLKKKVMYYTNISLFSGQAANWRDTFGFAVAPDPPKPDEIPL 543
Query: 178 ICRDIVIEYSQKIKNLGFTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELT 237
+CRDIV+EYS+KIK LGFTI ELLSE LGLNPSYLKE CAEGLF LGH+YP CPEP+LT
Sbjct: 544 VCRDIVMEYSKKIKELGFTILELLSEGLGLNPSYLKELNCAEGLFVLGHYYPPCPEPKLT 723
Query: 238 MGSTAHTDSTFMTLLLQDQLGGLQVLHEDKWV 269
MG+T HTD F+TLLLQDQLGGLQ+LHE++WV
Sbjct: 724 MGTTKHTDGNFITLLLQDQLGGLQILHENQWV 819
Score = 164 bits (416), Expect(2) = e-152
Identities = 79/104 (75%), Positives = 93/104 (88%)
Frame = +2
Query: 270 NVPPVHGALVVNIGDLLQLISNDKFVSVLHRVLSQNIGPRISVASFFLNSRDPIEGTSKI 329
+V PVHGALVVNIGDLLQLI+ND+FVSV HRVLSQNIGPRISVASFF+NS DPIEGT K+
Sbjct: 821 DVHPVHGALVVNIGDLLQLITNDRFVSVYHRVLSQNIGPRISVASFFVNSPDPIEGTLKV 1000
Query: 330 YGPIKELISEENPAIYKDITIKDFLAHFHTEGRNVKFLLEPFKL 373
YGPIKEL++EENP IY+D+TIKDFLAH++ +G + LEPF+L
Sbjct: 1001YGPIKELLTEENPPIYRDVTIKDFLAHYYAKGLDGNSSLEPFRL 1132
>TC14685 weakly similar to UP|Q43383 (Q43383) 2A6 protein, partial (38%)
Length = 1323
Score = 290 bits (741), Expect = 3e-79
Identities = 153/360 (42%), Positives = 221/360 (60%), Gaps = 4/360 (1%)
Frame = +1
Query: 17 YDREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISE---STASDSKLSIPIVDLK 73
+DR VK FD+SK GVKGL+DSG+ IP F +S+ SDSK IP +DL
Sbjct: 139 FDRAKAVKEFDESKTGVKGLIDSGIKTIPSFFVHPPETLSDLKPRPESDSKPEIPTIDLA 318
Query: 74 DIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQDADVRKEFY 133
++ R V+DQIR A GFFQV+NH +P+ +L + I FHEQ + R Y
Sbjct: 319 AVN---ESRAAVVDQIRRAASSVGFFQVVNHGVPLDLLRRTVAAIKAFHEQPEEERARVY 489
Query: 134 TRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEVPQICRDIVIEYSQKIKNL 193
TR++ VSY SNV LF + A+WRDT + P + +P++CR V+E+ +++ +
Sbjct: 490 TREMGTGVSYISNVDLFQSKAASWRDTLQLRMGPTAVDQSMIPEVCRQEVMEWDKEVVRV 669
Query: 194 GFTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGSTAHTDSTFMTLLL 253
G ++ LLSE LGL+ L E G +G +GH+YP CP+P+LT+G +H D +T++L
Sbjct: 670 GEVLYGLLSEGLGLDAGRLSEMGLVQGRVMVGHYYPFCPQPDLTVGLNSHADPGALTVVL 849
Query: 254 QDQLGGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHRVLSQNIG-PRISV 312
QD +GGLQV + WV+V P+ GALV+NIGDLLQ+ISN+++ S HRVL+ + PR+SV
Sbjct: 850 QDHIGGLQVRTTEGWVDVKPLDGALVINIGDLLQIISNEEYKSADHRVLANSANEPRVSV 1029
Query: 313 ASFFLNSRDPIEGTSKIYGPIKELISEENPAIYKDITIKDFLAHFHTEGRNVKFLLEPFK 372
A FLN + K++GP+ EL S E PA+Y++ T+ +FL F + + K L F+
Sbjct: 1030A-VFLNPGN----REKLFGPLPELTSAEKPALYRNFTLDEFLTRFFKKELDGKSLTNYFR 1194
>TC16060 weakly similar to UP|Q9SHK4 (Q9SHK4) F12K11.6, partial (8%)
Length = 835
Score = 275 bits (703), Expect = 9e-75
Identities = 135/222 (60%), Positives = 171/222 (76%)
Frame = +2
Query: 1 MVVENSNQLKESSDFTYDREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISESTA 60
M V+N +QL+E TYDREAEVKAFD +K+GVKGLV+SGV+KIPR+F + KL+I+E+
Sbjct: 173 MEVKNIHQLEEG---TYDREAEVKAFDATKLGVKGLVESGVTKIPRIFNSGKLDITENAP 343
Query: 61 SDSKLSIPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICR 120
+DS L++PI+DLKDIH NPA R VIDQIRSAC EWGFF V NH IP +L +MIDGI R
Sbjct: 344 TDSMLNVPIIDLKDIHNNPALRAAVIDQIRSACEEWGFFLVTNHGIPTTLLVDMIDGIRR 523
Query: 121 FHEQDADVRKEFYTRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEVPQICR 180
FHEQ +VRK++YTRDL K Y+SN L+ + +NWRDT G +AP P P+E+P + R
Sbjct: 524 FHEQVPEVRKQYYTRDLTSKTLYFSNASLYRDKYSNWRDTVGCLMAPKPPKPEELPHVFR 703
Query: 181 DIVIEYSQKIKNLGFTIFELLSEALGLNPSYLKEFGCAEGLF 222
DI+IEY ++K LG+ + ELLSEALGLNPS+LK+ C EGLF
Sbjct: 704 DIIIEYFNQVKALGYKMLELLSEALGLNPSFLKDLECGEGLF 829
>TC11427 weakly similar to UP|Q9LTH7 (Q9LTH7) Leucoanthocyanidin
dioxygenase-like protein (AT5g59540/f2o15_200), partial
(30%)
Length = 690
Score = 170 bits (431), Expect = 3e-43
Identities = 88/160 (55%), Positives = 115/160 (71%)
Frame = +1
Query: 214 EFGCAEGLFTLGHFYPTCPEPELTMGSTAHTDSTFMTLLLQDQLGGLQVLHEDKWVNVPP 273
EFG F LG P P + T MT+LLQDQ+GGLQ+LH+++WV+VPP
Sbjct: 1 EFGTRT--FILGPTIPHALNPN*LWARRSILILTLMTILLQDQIGGLQILHQNQWVDVPP 174
Query: 274 VHGALVVNIGDLLQLISNDKFVSVLHRVLSQNIGPRISVASFFLNSRDPIEGTSKIYGPI 333
VHG+LVVNIGDLLQL++N +F SV HRVLS+++GPRIS+ASFF+N +G SK+ GPI
Sbjct: 175 VHGSLVVNIGDLLQLMTNGQFSSVYHRVLSRHVGPRISIASFFVNISS--QGKSKVIGPI 348
Query: 334 KELISEENPAIYKDITIKDFLAHFHTEGRNVKFLLEPFKL 373
KEL+SEENP IY++ TIKD +AH+ +G + LEPFK+
Sbjct: 349 KELLSEENPPIYREATIKDVMAHYIEKGLDGNSSLEPFKM 468
>TC7931 weakly similar to UP|Q43640 (Q43640) Dioxygenase, partial (29%)
Length = 1477
Score = 160 bits (406), Expect = 2e-40
Identities = 103/300 (34%), Positives = 159/300 (52%), Gaps = 6/300 (2%)
Frame = +1
Query: 66 SIPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQD 125
++P+VDL R E + QI A E+GFFQVINH + ++++ ++ FH
Sbjct: 130 AVPVVDL-----GGHDRAETLKQIFKASEEYGFFQVINHGVSNELIEDTLNIFKEFHAMP 294
Query: 126 ADVRKEFYTRDLKKKVSYYSNVRLFNGQGAN-WRDTFGFAIAPDP----FNPDEVPQICR 180
A+ + +RD Y++ + N W+DT P F P + P R
Sbjct: 295 AEEKLSESSRDPNGSCMLYTSREINNKDCIQFWKDTLRHPCTPSEEFMEFWPLK-PARYR 471
Query: 181 DIVIEYSQKIKNLGFTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGS 240
++V +Y+++++ LG I E+L E LGL+P Y G +E L + YP CPEP LT+G
Sbjct: 472 EVVEKYTKELRELGSQILEMLCEGLGLDPRYCCA-GLSENPLLLSNHYPPCPEPSLTLGL 648
Query: 241 TAHTDSTFMTLLLQD-QLGGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLH 299
+ H D T +T+LLQD + LQV + +W+++ P+ A VVNIG LLQ+ISN + V V H
Sbjct: 649 SKHRDPTLVTILLQDTDVNALQVFKDGEWISIEPIPYAFVVNIGLLLQIISNGRLVGVEH 828
Query: 300 RVLSQNIGPRISVASFFLNSRDPIEGTSKIYGPIKELISEENPAIYKDITIKDFLAHFHT 359
RV++ + R +VA F ++ +I P K L+ IY+ I ++FL F T
Sbjct: 829 RVVTNSSISRTTVAYFIRPKKE------EIVEPAKALVRTGAHPIYRSIAFEEFLRIFKT 990
>TC8903 weakly similar to UP|Q9LSW7 (Q9LSW7)
1-aminocyclopropane-1-carboxylate oxidase
(AT5g43440/MWF20_15), partial (25%)
Length = 578
Score = 159 bits (402), Expect = 7e-40
Identities = 77/126 (61%), Positives = 96/126 (76%)
Frame = +2
Query: 248 FMTLLLQDQLGGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHRVLSQNIG 307
F T++LQDQ GGLQ LH D+W+NVP VHG LVVN GDLLQLI+NDKF+SV HRVL + G
Sbjct: 5 FFTIVLQDQQGGLQALHADQWINVPAVHGTLVVNTGDLLQLITNDKFLSVYHRVLVSHGG 184
Query: 308 PRISVASFFLNSRDPIEGTSKIYGPIKELISEENPAIYKDITIKDFLAHFHTEGRNVKFL 367
PR+S+ASFF+N EG K+YGPIKEL+SE NP +Y++ TI +FLAH +G +
Sbjct: 185 PRVSIASFFVNPVQE-EGAPKVYGPIKELLSEINPPVYRETTISEFLAHHFVKGLDGNSA 361
Query: 368 LEPFKL 373
L+PFKL
Sbjct: 362 LKPFKL 379
>TC14222 similar to UP|Q41681 (Q41681) 1-aminocyclopropane-1-carboxylate
oxidase homolog (ACC oxidase) , partial (97%)
Length = 1597
Score = 156 bits (395), Expect = 5e-39
Identities = 100/298 (33%), Positives = 165/298 (54%), Gaps = 9/298 (3%)
Frame = +1
Query: 66 SIPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQD 125
+ P+VD+ + N +RV ++ I+ AC WGFF+++NHEI +D ++ + + H
Sbjct: 52 NFPVVDMSKL--NTEERVATMELIKDACENWGFFELVNHEISTEFMDT-VERLTKEHY-- 216
Query: 126 ADVRKEFYTRDLKKKVSYYSNVRLFNG-QGANWRDTFGFAIAPDPFNPDEVPQICRD--- 181
K F + K+ V+ + + +W TF P N EVP + D
Sbjct: 217 ----KRFMEQRFKEMVATKGLETVQSEIDDLDWESTFFLRHLPSS-NISEVPDLDEDYRK 381
Query: 182 IVIEYSQKIKNLGFTIFELLSEALGLNPSYLKE--FGCAEGLF-TLGHFYPTCPEPELTM 238
++ E++ K++ L + +LL E LGL YLK+ +G F T YP CP+P+L
Sbjct: 382 VMKEFAVKLEKLAENLLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIK 561
Query: 239 GSTAHTDSTFMTLLLQD-QLGGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSV 297
G AHTD+ + LL QD ++ GLQ+L +D+W++VPP+ ++VVN+GD L++I+N K+ SV
Sbjct: 562 GLRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMRHSIVVNLGDQLEVITNGKYKSV 741
Query: 298 LHRVLSQNIGPRISVASFFLNSRDPIEGTSKIYGPIKELISE-ENPAIYKDITIKDFL 354
+HRV++Q G R+S+ASF+ +P G + P L+ E E +Y +D++
Sbjct: 742 MHRVIAQTDGARMSLASFY----NP--GDDAVISPAPSLVKEDETSQVYPKFVFEDYM 897
>TC14383 homologue to UP|Q8W3Y3 (Q8W3Y3) 1-aminocyclopropane-1-carboxylic
acid oxidase, complete
Length = 1249
Score = 149 bits (375), Expect = 1e-36
Identities = 99/313 (31%), Positives = 171/313 (54%), Gaps = 10/313 (3%)
Frame = +3
Query: 52 KLEISESTASDSKLSIPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVL 111
K ++ + S + P++ L+ + N +R ++ QI+ AC WGFF+++NH IP V+
Sbjct: 3 KEQVEKKIIS*EMANFPVISLEKL--NGEERKDIKLQIKDACENWGFFELVNHGIPHDVM 176
Query: 112 DEMIDGICRFHEQDADVRKEFYTRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFN 171
D ++ + + H + ++ K + + V+ +W TF P+ N
Sbjct: 177 DT-VERLTKEHYRICMEQRFKELMASKGLEAVQTEVK-----DMDWESTFHLRHLPES-N 335
Query: 172 PDEVPQIC---RDIVIEYSQKIKNLGFTIFELLSEALGLNPSYLKE-FGCAEG--LFTLG 225
E+P + R ++ +++ +I+ L + +LL E LGL YLK+ F + G T
Sbjct: 336 ISEIPDLTDEYRKVMKDFALRIEKLSEDLLDLLCENLGLEKGYLKKAFHGSRGPTFGTKV 515
Query: 226 HFYPTCPEPELTMGSTAHTDSTFMTLLLQD-QLGGLQVLHEDKWVNVPPVHGALVVNIGD 284
YP CP+P L G AHTD+ + LL QD ++ GLQ+L + +WV+VPP+ ++VVN+GD
Sbjct: 516 ANYPPCPKPNLVKGLRAHTDAGGIILLFQDDKVSGLQLLKDGEWVDVPPMRHSIVVNLGD 695
Query: 285 LLQLISNDKFVSVLHRVLSQNIGPRISVASFFLNSRDPIEGTSKIYGPIKELI---SEEN 341
L++I+N K+ SV HRV++Q G R+S+ASF+ +P G+ + P EL+ +EE
Sbjct: 696 QLEVITNGKYKSVEHRVIAQTDGTRMSIASFY----NP--GSDAVIYPAPELLEKEAEEK 857
Query: 342 PAIYKDITIKDFL 354
+Y +D++
Sbjct: 858 NQLYPKFVFEDYM 896
>TC18852 weakly similar to UP|Q9SHK4 (Q9SHK4) F12K11.6, partial (4%)
Length = 532
Score = 146 bits (368), Expect = 6e-36
Identities = 69/147 (46%), Positives = 106/147 (71%), Gaps = 3/147 (2%)
Frame = +1
Query: 18 DREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISESTASDSKLS---IPIVDLKD 74
++E ++KAFDD+K GVKGLVD+G++ IP++F + S+++AS S S IPI+DL++
Sbjct: 88 EQEQQLKAFDDTKAGVKGLVDAGITTIPQIFIAPPTDGSKTSASPSTHSHFQIPIIDLEN 267
Query: 75 IHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQDADVRKEFYT 134
+ + A+R +++ +++ A GFFQV+NH IP +L EMI+G+CRFHEQ DV+KEF+
Sbjct: 268 LQGHVAERKDIVKKVQVASETCGFFQVVNHGIPKEILHEMIEGVCRFHEQPLDVKKEFFN 447
Query: 135 RDLKKKVSYYSNVRLFNGQGANWRDTF 161
RD KK+ + SN L+ + ANWRD+F
Sbjct: 448 RDFSKKMRFNSNFDLYQSKVANWRDSF 528
>BI419185
Length = 433
Score = 132 bits (332), Expect = 9e-32
Identities = 64/139 (46%), Positives = 94/139 (67%)
Frame = +2
Query: 21 AEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISESTASDSKLSIPIVDLKDIHINPA 80
+E +AFD++K GVKGLVD+GV KIP +F+ + + ++T + + IP++DL +I P+
Sbjct: 20 SERRAFDETKAGVKGLVDAGVKKIPTLFHHQPDKFDKATNLGNHV-IPVIDLANIDKGPS 196
Query: 81 QRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQDADVRKEFYTRDLKKK 140
R +++ +R A WGFFQV+NH P+ VL+E+ +G+ RF EQDA+V KEFYTRD K
Sbjct: 197 LRQDIVHMLREASETWGFFQVVNHGTPLSVLEEIKNGVKRFFEQDAEVIKEFYTRDKFKS 376
Query: 141 VSYYSNVRLFNGQGANWRD 159
Y SN L++ NWRD
Sbjct: 377 FIYNSNFDLYSSPALNWRD 433
>TC15359 weakly similar to UP|Q84MB3 (Q84MB3) At1g06620, partial (27%)
Length = 575
Score = 131 bits (329), Expect = 2e-31
Identities = 59/101 (58%), Positives = 82/101 (80%)
Frame = +2
Query: 261 QVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHRVLSQNIGPRISVASFFLNSR 320
QVL++++WV+V PV GALV+N+GD+LQLISNDKF S HRVL+ +GPRISVA FF
Sbjct: 2 QVLYQNQWVDVKPVDGALVINLGDILQLISNDKFKSARHRVLANKVGPRISVACFFSTHF 181
Query: 321 DPIEGTSKIYGPIKELISEENPAIYKDITIKDFLAHFHTEG 361
P +++YGPIKEL+SEENP +YK+ T++D++AH+H++G
Sbjct: 182 QP---CNRVYGPIKELLSEENPPLYKETTVRDYVAHYHSKG 295
>TC7936 similar to UP|Q9SP53 (Q9SP53) Flavanone 3-hydroxylase, partial
(94%)
Length = 1492
Score = 130 bits (328), Expect = 3e-31
Identities = 80/255 (31%), Positives = 128/255 (49%), Gaps = 6/255 (2%)
Frame = +3
Query: 67 IPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQDA 126
IP++ L I +R E+ ++I AC WG FQV++H + ++ M F
Sbjct: 261 IPVISLAGIDEVDDRRSEICNKIVEACENWGIFQVVDHGVDTELVSHMTTLAKEFFALPP 440
Query: 127 DVRKEFYTRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAP----DPFNPDEVPQICRDI 182
+ + F KK + + L +WR+ + P D + P + +
Sbjct: 441 EEKLRFDMTG-GKKGGFIVSSHLQGESVQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAV 617
Query: 183 VIEYSQKIKNLGFTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGSTA 242
EYS+K+ L + E+LSEA+GL L + + ++YP CP+P+LT+G
Sbjct: 618 TEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDMDQKVVVNYYPKCPQPDLTLGLKR 797
Query: 243 HTDSTFMTLLLQDQLGGLQVLHED--KWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHR 300
HTD +TLLLQDQ+GGLQ ++ W+ V PV GA VVN+GD +SN +F + H+
Sbjct: 798 HTDPGTITLLLQDQVGGLQATRDNGKTWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQ 977
Query: 301 VLSQNIGPRISVASF 315
+ + R+S+A+F
Sbjct: 978 AVVNSNSSRLSIATF 1022
>TC8129 similar to UP|Q9XG83 (Q9XG83) GA 2-oxidase, partial (90%)
Length = 1278
Score = 116 bits (290), Expect = 7e-27
Identities = 87/301 (28%), Positives = 147/301 (47%), Gaps = 18/301 (5%)
Frame = +2
Query: 67 IPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQDA 126
+P VDL +P + +++ AC E+GFF++INH+IP+ ++ + + F Q
Sbjct: 197 VPEVDLS----HPEAKTLIVN----ACKEFGFFKIINHDIPMELISNLENEALSFFMQPQ 352
Query: 127 DVRKEFYTRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEV------PQICR 180
+++ D Y + R+ W + F PD +P+ + +
Sbjct: 353 SEKEKASLDD----PFGYGSKRIGKNGDVGWVEYLLFNTNPDVISPNPLLLLEQNQNVFS 520
Query: 181 DIVIEYSQKIKNLGFTIFELLSEALGLNP----SYLKEFGCAEGLFTLGHFYPTCPEPEL 236
V +Y+ +KNL I EL+++ LG+ S L ++ +F L H YP C E EL
Sbjct: 521 SAVNDYTLAVKNLSCEILELMADGLGIEQRNVFSRLIRDEKSDCIFRLNH-YPACGELEL 697
Query: 237 T-------MGSTAHTDSTFMTLLLQDQLGGLQV-LHEDKWVNVPPVHGALVVNIGDLLQL 288
+G HTD +++L + GLQ+ L + WV++PP + +N+GD LQ+
Sbjct: 698 QAIGGRNLIGFGEHTDPQMISVLRSNNTSGLQICLRDGTWVSIPPDQTSFFINVGDSLQV 877
Query: 289 ISNDKFVSVLHRVLSQNIGPRISVASFFLNSRDPIEGTSKIYGPIKELISEENPAIYKDI 348
++N F SV HRVLS R+S+ F P+ T K+ P+ L+S+E ++YK+
Sbjct: 878 MTNGSFRSVKHRVLSDTAMSRVSMIYF---GGAPL--TEKL-APLPSLVSKEEDSLYKEF 1039
Query: 349 T 349
T
Sbjct: 1040T 1042
>TC19303 similar to UP|Q84MB3 (Q84MB3) At1g06620, partial (15%)
Length = 472
Score = 106 bits (264), Expect = 7e-24
Identities = 55/89 (61%), Positives = 66/89 (73%)
Frame = +3
Query: 285 LLQLISNDKFVSVLHRVLSQNIGPRISVASFFLNSRDPIEGTSKIYGPIKELISEENPAI 344
LLQLI+ND F+SV HRVLS + GPR+S ASFFL D EG SK+YGPIKEL+SEENP I
Sbjct: 48 LLQLITNDIFISVYHRVLSSHRGPRVSAASFFLQY-DSDEGKSKVYGPIKELLSEENPPI 224
Query: 345 YKDITIKDFLAHFHTEGRNVKFLLEPFKL 373
YKD TI D +AH ++G + L PF+L
Sbjct: 225 YKDTTIADMMAHHLSKGLDGNSALHPFRL 311
>TC11971 weakly similar to UP|Q43419 (Q43419) ACC oxidase, partial (69%)
Length = 730
Score = 87.0 bits (214), Expect(2) = 1e-22
Identities = 51/133 (38%), Positives = 79/133 (59%), Gaps = 5/133 (3%)
Frame = +3
Query: 180 RDIVIEYSQKIKNLGFTIFELLSEALGLNPSYLKE-FGCAEG--LFTLGHFYPTCPEPEL 236
R + +Y +K+ L + L+SE LGL Y+K+ F ++G + T YP CP PEL
Sbjct: 327 RQTMDQYIEKLIQLAEDLSGLMSENLGLEKDYIKKAFSGSDGPAMGTKVAKYPQCPNPEL 506
Query: 237 TMGSTAHTDSTFMTLLLQ-DQLGGLQVLHEDKWVNVPP-VHGALVVNIGDLLQLISNDKF 294
G HTD+ + LLLQ DQ+ GL+ + KWV +PP + A+ +N GD ++++SN +
Sbjct: 507 VRGLREHTDAGGIILLLQDDQVPGLEFSKDGKWVEIPPSKNNAIFINTGDQIEVLSNGLY 686
Query: 295 VSVLHRVLSQNIG 307
SV+HRV++ G
Sbjct: 687 KSVVHRVMADKNG 725
Score = 35.8 bits (81), Expect(2) = 1e-22
Identities = 20/77 (25%), Positives = 41/77 (52%)
Frame = +2
Query: 65 LSIPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQ 124
++IPI+D + N +R E + + AC WG F + NH I +++++ I +E+
Sbjct: 2 MAIPIIDFSTL--NGDKRGETMALLHEACKNWGCFLIENHGIDKSLMEKVKQLINTHYEE 175
Query: 125 DADVRKEFYTRDLKKKV 141
++++ FY + K +
Sbjct: 176 --NLKQSFYESEKAKSL 220
>TC15818 similar to UP|Q9FFF6 (Q9FFF6) Leucoanthocyanidin dioxygenase-like
protein (AT5g05600/MOP10_14), partial (37%)
Length = 618
Score = 100 bits (250), Expect = 3e-22
Identities = 51/135 (37%), Positives = 81/135 (59%), Gaps = 1/135 (0%)
Frame = +2
Query: 228 YPTCPEPELTMGSTAHTDSTFMTLLLQDQ-LGGLQVLHEDKWVNVPPVHGALVVNIGDLL 286
YP CP+P+LT+G + H+D +T+LL D + GLQV + W+ V PV A ++NIGD +
Sbjct: 2 YPKCPQPDLTLGLSPHSDPGGLTILLPDDFVSGLQVRKGNYWITVKPVPNAFIINIGDQI 181
Query: 287 QLISNDKFVSVLHRVLSQNIGPRISVASFFLNSRDPIEGTSKIYGPIKELISEENPAIYK 346
Q++SN + SV HRV+ + R+S+A F+ D + P KEL++EE PA+Y
Sbjct: 182 QVLSNTIYKSVEHRVIVNSNQDRVSLAMFYNPKSD------LLIEPAKELVTEEKPALYP 343
Query: 347 DITIKDFLAHFHTEG 361
+T ++ + +G
Sbjct: 344 PMTYDEYRLYIRRQG 388
>BI420511
Length = 384
Score = 100 bits (248), Expect = 5e-22
Identities = 46/97 (47%), Positives = 72/97 (73%), Gaps = 1/97 (1%)
Frame = +3
Query: 228 YPTCPEPELTMGSTAHTDSTFMTLLLQD-QLGGLQVLHEDKWVNVPPVHGALVVNIGDLL 286
YP CP+PEL G AHTD+ + LLLQD ++ GLQ+L + +WV+VPP+ ++V+N+GD L
Sbjct: 36 YPACPKPELVKGLRAHTDAGGIILLLQDDKVSGLQLLKDGQWVDVPPMRHSIVINLGDQL 215
Query: 287 QLISNDKFVSVLHRVLSQNIGPRISVASFFLNSRDPI 323
++I+N K+ SV HRV+++ G R+S+ASF+ + D +
Sbjct: 216 EVITNGKYKSVEHRVIAKTDGTRMSIASFYNPANDAV 326
>AW719573
Length = 521
Score = 99.0 bits (245), Expect = 1e-21
Identities = 55/152 (36%), Positives = 90/152 (59%), Gaps = 2/152 (1%)
Frame = +2
Query: 167 PDPFNPDEVPQICRDIVIEYSQKIKNLGFTIFELLSEALGLNPSYLKEFGCAEGLFTLG- 225
P FN +PQ R+ + Y ++ NL I EL++ AL ++P + E EG T+
Sbjct: 32 PHLFN--HIPQPFRENLEIYCAELNNLSIKIMELMANALKIDPKEITEI-YNEGSQTIRM 202
Query: 226 HFYPTCPEPELTMGSTAHTDSTFMTLLLQ-DQLGGLQVLHEDKWVNVPPVHGALVVNIGD 284
++YP CP+PE +G +HTD +T+LLQ + + GLQ+ + WV V P+ A ++NIGD
Sbjct: 203 NYYPPCPQPERVIGLKSHTDGAGLTILLQANDIEGLQIRKDGHWVPVQPLPNAFIINIGD 382
Query: 285 LLQLISNDKFVSVLHRVLSQNIGPRISVASFF 316
+L++ +N + S+ HR + RIS+A+F+
Sbjct: 383 MLEITTNGIYASIEHRATVNSEKERISLATFY 478
>BE122499
Length = 321
Score = 97.1 bits (240), Expect = 4e-21
Identities = 46/100 (46%), Positives = 71/100 (71%)
Frame = +2
Query: 21 AEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISESTASDSKLSIPIVDLKDIHINPA 80
+E KAFD++K GVKGLVD+GV KIP F+ + + +++ S + +P++DL +I +P+
Sbjct: 23 SERKAFDETKAGVKGLVDAGVKKIPTFFHHQHDKFEQASNSGDHV-VPVIDLANIDKDPS 199
Query: 81 QRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICR 120
R EV++Q+R A WGFFQ +NH IP+ VL+E+ DG+ R
Sbjct: 200 LRQEVVNQLREASETWGFFQSVNHGIPLSVLEEIQDGVQR 319
>TC10874 similar to UP|Q8LF06 (Q8LF06) Leucoanthocyanidin dioxygenase-like
protein, partial (38%)
Length = 723
Score = 90.5 bits (223), Expect = 4e-19
Identities = 53/139 (38%), Positives = 79/139 (56%), Gaps = 2/139 (1%)
Frame = +2
Query: 236 LTMGSTAHTDSTFMTLLL-QDQLGGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKF 294
LT+G ++H+D MT+LL DQ+ GLQV D W+ V P A +VNIGD +Q++SN +
Sbjct: 2 LTLGLSSHSDPGGMTMLLPDDQVRGLQVRKGDDWITVNPARHAFIVNIGDQIQVLSNAIY 181
Query: 295 VSVLHRVLSQNIGPRISVASFFLNSRD-PIEGTSKIYGPIKELISEENPAIYKDITIKDF 353
SV HRV+ + R+S+A F+ D PIE P+KEL++ + PA+Y +T ++
Sbjct: 182 TSVEHRVIVNSDKERVSLAFFYNPKSDIPIE-------PVKELVTPDKPALYPAMTFDEY 340
Query: 354 LAHFHTEGRNVKFLLEPFK 372
G K +E K
Sbjct: 341 RLFIRMRGPRGKSQVESMK 397
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.140 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,056,081
Number of Sequences: 28460
Number of extensions: 81590
Number of successful extensions: 491
Number of sequences better than 10.0: 120
Number of HSP's better than 10.0 without gapping: 436
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 438
length of query: 373
length of database: 4,897,600
effective HSP length: 92
effective length of query: 281
effective length of database: 2,279,280
effective search space: 640477680
effective search space used: 640477680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC139601.6


