

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139601.1 - phase: 0 /pseudo
(2139 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
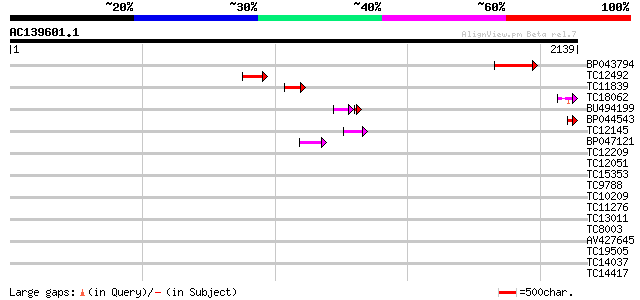
Score E
Sequences producing significant alignments: (bits) Value
BP043794 211 7e-55
TC12492 weakly similar to UP|AAS53933 (AAS53933) AFR562Cp, parti... 128 1e-29
TC11839 similar to UP|STH1_YEAST (P32597) Nuclear protein STH1/N... 79 1e-14
TC18062 similar to UP|Q7Y244 (Q7Y244) Fiber protein Fb22 (Fragme... 77 4e-14
BU494199 51 4e-10
BP044543 57 3e-08
TC12145 similar to UP|Q9ZVP6 (Q9ZVP6) T7A14.1 protein (Fragment)... 57 3e-08
BP047121 43 6e-04
TC12209 similar to UP|O60177 (O60177) DEAD box helicase, partial... 42 0.001
TC12051 34 0.21
TC15353 weakly similar to UP|AAQ81937 (AAQ81937) SCC3, partial (7%) 34 0.27
TC9788 similar to PIR|T47605|T47605 RING finger-like protein - A... 34 0.27
TC10209 similar to UP|Q9SJQ5 (Q9SJQ5) At2g36500 protein, partial... 33 0.46
TC11276 weakly similar to UP|HDA1_CHICK (P56517) Histone deacety... 32 1.0
TC13011 similar to UP|Q9JM93 (Q9JM93) SRp25 nuclear protein, par... 32 1.0
TC8003 homologue to PIR|S52023|S52023 translation initiation fac... 32 1.0
AV427645 32 1.4
TC19505 weakly similar to UP|Q9FG35 (Q9FG35) Emb|CAB82953.1, par... 31 1.8
TC14037 similar to UP|O81698 (O81698) Proline-rich protein, part... 31 1.8
TC14417 homologue to UP|AAR23806 (AAR23806) Initiation factor eI... 31 2.3
>BP043794
Length = 490
Score = 211 bits (538), Expect = 7e-55
Identities = 110/161 (68%), Positives = 125/161 (77%)
Frame = +2
Query: 1829 NLRGSSAHVTNMTEIIQRRCKSVISKLQRRIDKEGHQIVPLLTDLWKRIENSGFAGGSGN 1888
NL GSS T MTEIIQR CK+VISKLQRRIDK+G Q+VPLLT KRIENSG+ GSG
Sbjct: 2 NLSGSSTRTTKMTEIIQRGCKNVISKLQRRIDKDGQQVVPLLTAFLKRIENSGYTAGSGK 181
Query: 1889 NLLDLRKIDQRINRLEYSGVMEFVFDVQFMLKSAMQFYGYSYEVRTEARKVHDLFFDILK 1948
LLDLRKIDQRI+RLEY+GVME V DVQFML+SAM YGYS EV EA +VH+LFFDILK
Sbjct: 182 TLLDLRKIDQRIDRLEYNGVMELVSDVQFMLRSAMHSYGYSEEVNNEADRVHNLFFDILK 361
Query: 1949 TTFSDIDFGEAKSALSFTSQISANAGASSKQATVFPSKRKR 1989
F ++DF EA++ LS Q+SA+A AS Q TV PSKR R
Sbjct: 362 IAFPNVDFREARNGLSSPPQVSASAVASPXQVTVGPSKRNR 484
>TC12492 weakly similar to UP|AAS53933 (AAS53933) AFR562Cp, partial (7%)
Length = 922
Score = 128 bits (321), Expect = 1e-29
Identities = 59/94 (62%), Positives = 74/94 (77%)
Frame = +1
Query: 877 KYYNLAHAVNEKVLRQPSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQV 936
+Y + H++ EKV QPS+L+ G LR YQ+ GLQWMLSL+NN LNGILADEMGLGKT+Q
Sbjct: 586 QYNSAIHSIQEKVTEQPSILQGGELRSYQIEGLQWMLSLFNNNLNGILADEMGLGKTIQT 765
Query: 937 MALIAYLMEFKGNYGPHLIIVPNAVLVNWKCAFN 970
++LIA+LME+KG GPHLI+ P AVL NW F+
Sbjct: 766 ISLIAHLMEYKGVTGPHLIVAPKAVLPNWMNEFS 867
>TC11839 similar to UP|STH1_YEAST (P32597) Nuclear protein STH1/NPS1, partial
(4%)
Length = 519
Score = 78.6 bits (192), Expect = 1e-14
Identities = 37/79 (46%), Positives = 56/79 (70%)
Frame = +2
Query: 1038 QNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLH 1097
+N L+ELW+LLN LLP +F++ + F+ WF+KPF+ N +A+ L E+ ++II+RLH
Sbjct: 212 KNSLEELWALLNFLLPNIFNSSEDFSQWFNKPFESSGDN-SADEALLSEEENLLIINRLH 388
Query: 1098 QILEPFMLRRRVEEVEGSL 1116
Q+L PF+LRR +V SL
Sbjct: 389 QVLRPFVLRRLKHKVLISL 445
>TC18062 similar to UP|Q7Y244 (Q7Y244) Fiber protein Fb22 (Fragment), partial
(29%)
Length = 621
Score = 76.6 bits (187), Expect = 4e-14
Identities = 45/88 (51%), Positives = 52/88 (58%), Gaps = 15/88 (17%)
Frame = -3
Query: 2067 KSSVKHRIGSAGPVSPPKIVVHTVLAERSPTPGSGSTPR-----------AGH----AHT 2111
KS VK R GS+GPVSPP + +P SGS PR AG +
Sbjct: 619 KSLVKTRTGSSGPVSPPS------MGPAIKSPRSGSNPRDSRPVQQAAQAAGRVGQPSQQ 458
Query: 2112 SNGSGGSVGWANPVKRMRTDSGKRRPSH 2139
NGSGGS+GWANPVKR+R+DSGKRRPSH
Sbjct: 457 PNGSGGSLGWANPVKRLRSDSGKRRPSH 374
>BU494199
Length = 570
Score = 50.8 bits (120), Expect(2) = 4e-10
Identities = 33/80 (41%), Positives = 44/80 (54%), Gaps = 4/80 (5%)
Frame = +1
Query: 1221 HRVLLFSTMTKLLDILEEYL---QWRRLVYRRIDGTTALEDRESAIVDFNS-PNSDCFIF 1276
HRVL+F+ LDI+E L + + Y R+DG+ E R + FNS P D +
Sbjct: 190 HRVLIFAQHKAFLDIIERDLFQTHMKNVTYLRLDGSVEPEKRFEIVKAFNSDPTID--VL 363
Query: 1277 LLSIRAAGRGLNLQSADTVV 1296
LL+ G GLNL SADT+V
Sbjct: 364 LLTTHVGGLGLNLTSADTLV 423
Score = 32.3 bits (72), Expect(2) = 4e-10
Identities = 14/25 (56%), Positives = 18/25 (72%)
Frame = +2
Query: 1301 DPNPKNEEQAVARAHRIGQKREVKV 1325
D NP + QA+ RAHR+GQK+ V V
Sbjct: 437 DWNPMRDHQAMDRAHRLGQKKVVNV 511
>BP044543
Length = 418
Score = 57.0 bits (136), Expect = 3e-08
Identities = 25/34 (73%), Positives = 26/34 (75%)
Frame = -3
Query: 2106 AGHAHTSNGSGGSVGWANPVKRMRTDSGKRRPSH 2139
AG NGSGG VGW PVKR+RTDSGKRRPSH
Sbjct: 347 AGRQSQPNGSGGPVGWQTPVKRLRTDSGKRRPSH 246
>TC12145 similar to UP|Q9ZVP6 (Q9ZVP6) T7A14.1 protein (Fragment), partial
(76%)
Length = 664
Score = 57.0 bits (136), Expect = 3e-08
Identities = 40/93 (43%), Positives = 51/93 (54%), Gaps = 3/93 (3%)
Frame = +2
Query: 1261 SAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQK 1320
+AI FN + DC FLLS++AAG LNL A V + +P NP E+QA R HRIGQ
Sbjct: 8 AAIKRFND-DPDCKFFLLSLKAAGVALNLTVASHVFLMEPWWNPDAEQQAQDRIHRIGQN 184
Query: 1321 ---REVKVIYMEAVVDKISSHQKEDEMRIGGTI 1350
R VK I V ++I Q++ E G I
Sbjct: 185 KPIRVVKFITENTVEERILELQQKKEAVSEGLI 283
>BP047121
Length = 525
Score = 42.7 bits (99), Expect = 6e-04
Identities = 29/104 (27%), Positives = 54/104 (51%), Gaps = 5/104 (4%)
Frame = -1
Query: 1095 RLHQILEPFMLRRRVEEVEGSLPPKVS-----IVLRCRMSAFQSAIYDWIKSTGTLRLNP 1149
R+ IL PF+LRR +V L PK+ ++ + + SA++ AI ++ ++ R+
Sbjct: 525 RMKSILGPFILRRLKSDVMQQLVPKIQQVEYVVMEKQQESAYKEAIEEY-RAVSQARM-- 355
Query: 1150 EEEQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHPLLNYPFFSD 1193
+ S + + + + + +NN ++ RK NHPLL +SD
Sbjct: 354 -AKCSELNSNNVLEVLPRRQINNYFVQFRKIANHPLLIRRIYSD 226
>TC12209 similar to UP|O60177 (O60177) DEAD box helicase, partial (6%)
Length = 550
Score = 41.6 bits (96), Expect = 0.001
Identities = 25/80 (31%), Positives = 43/80 (53%)
Frame = +1
Query: 1003 IIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAF 1062
I+ Q +K++ + AR + R LTGTP+ N++ EL+SL++ L + ++ K F
Sbjct: 289 ILDEAQCIKNKSTGAARACCTLQAIYRFCLTGTPMMNNVGELYSLIHFLRIKPYNEWKRF 468
Query: 1063 NDWFSKPFQKEDPNQNAEND 1082
D F K DP+++ D
Sbjct: 469 QDSFG-CLTKPDPSRSRRRD 525
>TC12051
Length = 629
Score = 34.3 bits (77), Expect = 0.21
Identities = 25/88 (28%), Positives = 42/88 (47%), Gaps = 2/88 (2%)
Frame = +3
Query: 1698 EGEEEQVLQKPKIKRK--RSLRVRPRHTMEKPEDKSGSEMASLQRGQSFLLPDKKYPLQS 1755
E + V ++ K +RK +++ RP +T EKP+D SG++ + LPD +Q
Sbjct: 159 ENKARVVREEKKRRRKWAQNITPRPSNTREKPQDISGTKPQQEPNTAAGFLPDN--IVQM 332
Query: 1756 RINQESKTFGDSSSNKHDKNEPILKNKR 1783
QE + F S + D +P K+
Sbjct: 333 LAAQEKQVFLSDSDEEKDGVKPTNSKKK 416
>TC15353 weakly similar to UP|AAQ81937 (AAQ81937) SCC3, partial (7%)
Length = 528
Score = 33.9 bits (76), Expect = 0.27
Identities = 21/54 (38%), Positives = 28/54 (50%), Gaps = 1/54 (1%)
Frame = +1
Query: 1496 DSTEIGSERRRGRPKGKKN-PSYKELEDSSEEISEDRNEDSAHDEGEIGEFEDD 1548
D E S RRRGRP+ ++N P+ K +D S ED S D G E ++D
Sbjct: 64 DEKEGVSIRRRGRPRKRQNIPAKKLFDDQSSSEDEDSISASEQDGGRQEEDDED 225
>TC9788 similar to PIR|T47605|T47605 RING finger-like protein - Arabidopsis
thaliana {Arabidopsis thaliana;}, partial (6%)
Length = 696
Score = 33.9 bits (76), Expect = 0.27
Identities = 27/97 (27%), Positives = 47/97 (47%), Gaps = 7/97 (7%)
Frame = +2
Query: 1270 NSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKR--EVKVIY 1327
+S C + L+ AA L+L V + +P + EEQ ++RAHR+G R V+ +
Sbjct: 20 DSSCMVLLMDGSAA-LVLDLSFVSHVFLMEPIWDRSLEEQVISRAHRMGASRPIHVETLA 196
Query: 1328 MEAVVDK--ISSHQKEDEMR---IGGTIDMEDELAGK 1359
M +++ + Q D+ R I + ED+ G+
Sbjct: 197 MRGTIEEQMLEFLQDADKCRRSPIKDAVKSEDDSGGR 307
>TC10209 similar to UP|Q9SJQ5 (Q9SJQ5) At2g36500 protein, partial (26%)
Length = 659
Score = 33.1 bits (74), Expect = 0.46
Identities = 20/50 (40%), Positives = 31/50 (62%), Gaps = 2/50 (4%)
Frame = +1
Query: 308 QQSSANKSLGADSSLNAKASSSRSDPEPAKMQYVR--QLSQHASLDGGST 355
QQ S+ KSL A++ NAKASS S P+ + V+ +LS+ ++ G+T
Sbjct: 178 QQRSSKKSLPAENGTNAKASSPPSQPDEGGERTVKKLRLSKALTIPEGTT 327
>TC11276 weakly similar to UP|HDA1_CHICK (P56517) Histone deacetylase 1 (HD1),
partial (5%)
Length = 636
Score = 32.0 bits (71), Expect = 1.0
Identities = 17/57 (29%), Positives = 30/57 (51%)
Frame = +3
Query: 1410 DEERCQETVHDVPSLQEVNRMIARNEEEVELFDQMDEEEDWLEEMTRYDQVPDWIRA 1466
+EE+ E + P +E + + EEE + D+ EEE E+ + D V +W++A
Sbjct: 69 EEEKKDEPKSEEPQKEEAKKEEPKKEEEKK--DEKKEEEKKKEQPAQIDPVLEWVKA 233
>TC13011 similar to UP|Q9JM93 (Q9JM93) SRp25 nuclear protein, partial (10%)
Length = 593
Score = 32.0 bits (71), Expect = 1.0
Identities = 40/158 (25%), Positives = 59/158 (37%), Gaps = 1/158 (0%)
Frame = -3
Query: 1480 KRPSKKNALSGGNVVLDSTEIGSERRRGRPKGKKNPSYKELEDSSEEISEDRNEDSAHDE 1539
K+P+KK A ++G KK KE DS EE ED E+S H E
Sbjct: 585 KKPAKKGA-----------------KKGADHSKKRKKAKEPSDSEEEEEEDA-EESDHSE 460
Query: 1540 GEI-GEFEDDGYSGAGIAQPVDKDKLDDVTPSDAEYECPRSSSESARNNNVVEGGSSASS 1598
E+ GE E G T + P+ S + + GG++A++
Sbjct: 459 DEVNGEAEVKRSRGG-------------KTSGKGRGKAPKKSGAEVKGSGRGRGGAAAAA 319
Query: 1599 AGVQRLTQAVSPSVSSQKFASLSALDAKPSSISKKMVH 1636
+ SP SS L L + P S+ ++H
Sbjct: 318 NKSGSSGKRGSPRKSS*LSFLLEILFSTPRE-SQHVIH 208
>TC8003 homologue to PIR|S52023|S52023 translation initiation factor
eIF-4A.14 - common tobacco {Nicotiana tabacum;} , partial
(34%)
Length = 708
Score = 32.0 bits (71), Expect = 1.0
Identities = 22/97 (22%), Positives = 43/97 (43%)
Frame = +1
Query: 1224 LLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAA 1283
++F + +D L + ++ R G R+ + +F S +S L++
Sbjct: 25 VIFVNTRRKVDWLTDKMRSRDHTVSATHGDMDQNTRDIIMREFRSGSSRV---LITTDLL 195
Query: 1284 GRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQK 1320
RG+++Q V+ YD P+N + R+ R G+K
Sbjct: 196 ARGIDVQQVSLVINYDLPTQPENYLHRIGRSGRFGRK 306
>AV427645
Length = 403
Score = 31.6 bits (70), Expect = 1.4
Identities = 19/56 (33%), Positives = 24/56 (41%), Gaps = 7/56 (12%)
Frame = +1
Query: 1532 NEDSAHDEGEIGEFEDDGYSGAGIAQPVD-----KDKLDDVTPSD--AEYECPRSS 1580
+ED HD ++ F+DD YSG P+D D DD E CP S
Sbjct: 202 SEDDMHDANDVESFDDDFYSGDAEEAPLDYYSDYDDDADDYFDDSDRIESRCPEVS 369
>TC19505 weakly similar to UP|Q9FG35 (Q9FG35) Emb|CAB82953.1, partial (12%)
Length = 723
Score = 31.2 bits (69), Expect = 1.8
Identities = 40/137 (29%), Positives = 52/137 (37%), Gaps = 17/137 (12%)
Frame = +1
Query: 315 SLGADSSLNAKASSSRSDPE-PAKMQYVRQLSQHASLDG------------GSTKEVGSG 361
S A SS ++ASS S+PE P ++ S SL GS
Sbjct: 265 SAAATSSPTSEASSQSSNPEKPLRLPMFLCSSSSLSLFS*LFPLLLTLHLPGSPTSSPPP 444
Query: 362 NYAKPQGGPSQMPQKLNGF----TKNQLHVLKAQILAFRRLKKGDGILPQELLEAISPPP 417
P P +P LN T + +L QIL RR D L LL + P P
Sbjct: 445 PPPPPPPPPPPVPLNLNCLPFSPTFSLTPLLHPQILHRRRATVLD--LAMLLLHLLIPKP 618
Query: 418 LDLHVQQPIHSAGAQNQ 434
H QQP+H ++Q
Sbjct: 619 AQKHWQQPLHKTKLRDQ 669
>TC14037 similar to UP|O81698 (O81698) Proline-rich protein, partial (25%)
Length = 465
Score = 31.2 bits (69), Expect = 1.8
Identities = 19/75 (25%), Positives = 35/75 (46%)
Frame = +2
Query: 298 GVNGMPSMHPQQSSANKSLGADSSLNAKASSSRSDPEPAKMQYVRQLSQHASLDGGSTKE 357
GVNG P + ++S+ + + S ++++S +DP P YV Q + + G+
Sbjct: 50 GVNGEPRRNVKESALRYHVSDEESHRTESTTSEADPHPE--PYVNQSAFPGTNYAGNGAI 223
Query: 358 VGSGNYAKPQGGPSQ 372
SG+ + G Q
Sbjct: 224 TSSGSKSSADGSTEQ 268
>TC14417 homologue to UP|AAR23806 (AAR23806) Initiation factor eIF4A-15,
complete
Length = 1563
Score = 30.8 bits (68), Expect = 2.3
Identities = 29/127 (22%), Positives = 53/127 (40%), Gaps = 2/127 (1%)
Frame = +2
Query: 1196 KDFMVKCCGKLWMLDRI--LIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGT 1253
K F V + W L+ + L + V+ +T K+ D L + ++ G
Sbjct: 848 KQFYVNVEKEEWKLETLCDLYETLAITQSVIFVNTRRKV-DWLTDKMRSNDHTVSATHGD 1024
Query: 1254 TALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVAR 1313
R+ + +F S +S L++ RG+++Q V+ YD P+N + R
Sbjct: 1025 MDQNTRDIIMREFRSGSSRV---LITTDLLARGIDVQQVSLVINYDLPTQPENYLHRIGR 1195
Query: 1314 AHRIGQK 1320
+ R G+K
Sbjct: 1196 SGRFGRK 1216
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.130 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,973,753
Number of Sequences: 28460
Number of extensions: 403913
Number of successful extensions: 1904
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 1853
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1892
length of query: 2139
length of database: 4,897,600
effective HSP length: 105
effective length of query: 2034
effective length of database: 1,909,300
effective search space: 3883516200
effective search space used: 3883516200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC139601.1


