

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139525.8 - phase: 0
(676 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
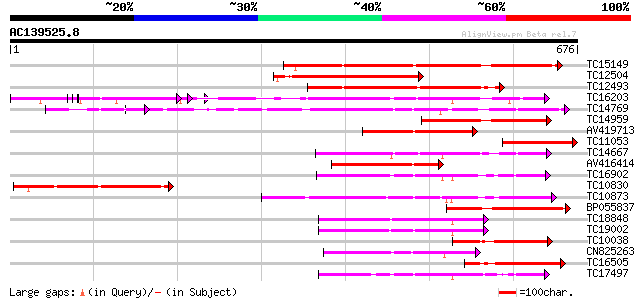
Score E
Sequences producing significant alignments: (bits) Value
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 321 3e-88
TC12504 weakly similar to UP|Q94JL9 (Q94JL9) At1g68400/T2E12_5, ... 315 1e-86
TC12493 similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (35%) 227 4e-60
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 198 3e-51
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 194 5e-50
TC14959 similar to GB|AAN64529.1|24797034|BT001138 At5g58299/At5... 155 3e-38
AV419713 154 3e-38
TC11053 weakly similar to UP|Q9FHK7 (Q9FHK7) Receptor-like prote... 152 2e-37
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 149 2e-36
AV416414 144 6e-35
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 142 2e-34
TC10830 weakly similar to UP|Q94JL9 (Q94JL9) At1g68400/T2E12_5, ... 142 2e-34
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 141 3e-34
BP055837 141 4e-34
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 140 5e-34
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 138 3e-33
TC10038 similar to UP|Q9FHK7 (Q9FHK7) Receptor-like protein kina... 136 1e-32
CN825263 127 8e-30
TC16505 similar to GB|AAN64529.1|24797034|BT001138 At5g58299/At5... 126 1e-29
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 125 3e-29
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 321 bits (822), Expect = 3e-88
Identities = 170/340 (50%), Positives = 233/340 (68%), Gaps = 7/340 (2%)
Frame = +3
Query: 327 NSNGGGGDSSDGT------SGTDMSKLVFF-DRRNGFELEDLLRASAEMLGKGSLGTVYR 379
N NG G ++ T +G KLVFF + GF+LEDLLRASAE+LGKG+ GT Y+
Sbjct: 147 NGNGNGYSAAAATINKVEANGVGAKKLVFFGNSARGFDLEDLLRASAEVLGKGTFGTAYK 326
Query: 380 AVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYL 439
AVL+ G VAVKRLKD + EF+ ++ +G + H ++V LRAYY++++EKLLVYDY+
Sbjct: 327 AVLETGLVVAVKRLKDVT-ISEKEFKDKIETVGAMDHQSLVPLRAYYFSRDEKLLVYDYM 503
Query: 440 SNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLL 499
GSL ALLHGN+G GR PL+W R + LGAARG+ +H++ V HGN+K+SN+LL
Sbjct: 504 PMGSLSALLHGNKGAGRTPLNWEIRSGIALGAARGIEYLHSQ--GPNVSHGNIKASNILL 677
Query: 500 DKNGVACISDFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGK 559
K+ A +SDFGL+ L+ P R+ GYRAPE T+ +++SQ+ADVYSFGVLLLE+LTGK
Sbjct: 678 TKSYEAKVSDFGLAHLVGPSSTPNRVAGYRAPEVTDPRKVSQKADVYSFGVLLLELLTGK 857
Query: 560 APSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSML 619
AP+ E VDLP+WV+S+VREEWT EVFD ELLRY+N+EEE+V +L
Sbjct: 858 APT-----------HALLNEEGVDLPRWVQSLVREEWTSEVFDLELLRYQNVEEEMVQLL 1004
Query: 620 HVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSPLCEDYDE 659
+ + C P++RP++ +V + IE++ +S L ED D+
Sbjct: 1005QLAVDCAAPYPDRRPSIAEVTRSIEELC--RSNLKEDQDQ 1118
>TC12504 weakly similar to UP|Q94JL9 (Q94JL9) At1g68400/T2E12_5, partial
(21%)
Length = 586
Score = 315 bits (808), Expect = 1e-86
Identities = 163/182 (89%), Positives = 167/182 (91%), Gaps = 3/182 (1%)
Frame = +1
Query: 315 KRKS---YGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRNGFELEDLLRASAEMLGK 371
KRKS YGSEKKVY S GDS DGTSGT+ SKLVFFDRR FELEDLLRASAEMLGK
Sbjct: 55 KRKSGSSYGSEKKVYAS----GDS-DGTSGTERSKLVFFDRRGEFELEDLLRASAEMLGK 219
Query: 372 GSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYAKEE 431
GSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHP++VKLRAYYYAKEE
Sbjct: 220 GSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPDMVKLRAYYYAKEE 399
Query: 432 KLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGN 491
KLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTR+SL LGAARGLARIH EYS AKVPHGN
Sbjct: 400 KLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRVSLALGAARGLARIHAEYSTAKVPHGN 579
Query: 492 VK 493
VK
Sbjct: 580 VK 585
>TC12493 similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (35%)
Length = 837
Score = 227 bits (579), Expect = 4e-60
Identities = 118/234 (50%), Positives = 162/234 (68%)
Frame = +3
Query: 356 FELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLK 415
F L++LLRASAE+LGKG+ GT Y+A ++ G +VAVKRLKD EF + ++ +GKL
Sbjct: 177 FSLDELLRASAEVLGKGTFGTTYKATMEMGRSVAVKRLKDVT-ATEMEFREKIEEVGKLV 353
Query: 416 HPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGL 475
H N+V LR YY++++EKL+VYDY+ GSL ALLH N G GR PL+W TR ++ LGAA G+
Sbjct: 354 HENLVPLRGYYFSRDEKLVVYDYMPMGSLSALLHANNGAGRTPLNWETRSAIALGAAHGI 533
Query: 476 ARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPEQTE 535
A +H++ + HGN+KSSN+LL K+ +SD GL+ L P R+ GYRAPE T+
Sbjct: 534 AYLHSQGPTSS--HGNIKSSNILLTKSFEPRVSDLGLAYLALPTSTPNRISGYRAPEVTD 707
Query: 536 QKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVR 589
+++SQ+ADVYSFG++LLE+LTGK P+ S N E VDLP+WV+
Sbjct: 708 ARKVSQKADVYSFGIMLLELLTGKPPT---HSSLN--------EEGVDLPRWVQ 836
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 198 bits (503), Expect = 3e-51
Identities = 170/594 (28%), Positives = 269/594 (44%), Gaps = 19/594 (3%)
Frame = +2
Query: 69 TLVLPSLNLRGPIDA-LSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSG 127
TL L + G I A + +L L+ L L N G + + L + ++GN+ +G
Sbjct: 1496 TLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFE-IPMLTKVNISGNNLTG 1672
Query: 128 QIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNL 187
IP I+ +L +DLS NNLAG++P + L +L L L N +SG +PD M +L
Sbjct: 1673 PIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSL 1852
Query: 188 TELNMTNNEFYGKVPN--TMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTV 245
T L++++N F G VP L D++F+GN LC
Sbjct: 1853 TTLDLSSNNFTGTVPTGGQFLVFNYDKTFAGNPNLCFP---------------------- 1966
Query: 246 PSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCARGRGVNS 305
+ +S P+ + +++ +++ I +A V L+ VT VV
Sbjct: 1967 --HRASCPSVLYDSLRKTRAKTARVRAIVIGIALATAVLLVAVTVHVVR----------- 2107
Query: 306 NSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRNGFELEDLLRAS 365
KR+ + ++ KL F R + ED++
Sbjct: 2108 ---------KRRLHRAQAW---------------------KLTAFQRLE-IKAEDVVECL 2194
Query: 366 AE--MLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCAR-HEFEQYMDVIGKLKHPNIVKL 422
E ++GKG G VYR + +G+ VA+KRL + F ++ +GK++H NI++L
Sbjct: 2195 KEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRL 2374
Query: 423 RAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEY 482
Y K+ LL+Y+Y+ NGSL LHG +G L W R + + AARGL +H +
Sbjct: 2375 LGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGG---HLRWEMRYKIAVEAARGLCYMHHDC 2545
Query: 483 SAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLG------GYRAPEQTEQ 536
S + H +VKS+N+LLD + A ++DFGL+ L A+ + GY APE
Sbjct: 2546 SPL-IIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYT 2722
Query: 537 KRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVRE-- 594
++ +++DVYSFGV+LLE++ G+ P V E VD+ WV + E
Sbjct: 2723 LKVDEKSDVYSFGVVLLELIIGRKP-------------VGEFGDGVDIVGWVNKTMSELS 2863
Query: 595 -----EWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMI 643
V D L Y ++ M ++ + CV + RPTM +VV M+
Sbjct: 2864 QPSDTALVLAVVDPRLSGYP--LTSVIHMFNIAMMCVKEMGPARPTMREVVHML 3019
Score = 88.2 bits (217), Expect = 4e-18
Identities = 47/123 (38%), Positives = 71/123 (57%)
Frame = +2
Query: 83 ALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLLRL 142
A S+ +LRLL++ N L G + SL N T L L++ N+ +G IPPE+SS+ +L+ L
Sbjct: 824 AFGSMENLRLLEMANCNLTGEIPPSL-GNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSL 1000
Query: 143 DLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVP 202
DLS N+L G+IP S+L NL + N G++P +PNL L + N F +P
Sbjct: 1001DLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLP 1180
Query: 203 NTM 205
+ +
Sbjct: 1181HNL 1189
Score = 75.5 bits (184), Expect = 3e-14
Identities = 71/269 (26%), Positives = 114/269 (41%), Gaps = 31/269 (11%)
Frame = +2
Query: 1 MNYVIMFFFFLFLSIYIVPCLTHNDTQALTLFRQQ---TDTHGQLLTNWTGPEACSA--S 55
++Y+++ F L + V + +D AL ++ L +W + SA S
Sbjct: 122 VSYLLVLCFTLIWFRWTVVYSSFSDLDALLKLKESMKGAKAKHHALEDWKFSTSLSAHCS 301
Query: 56 WHGVTCTPNNRVTTLVLPSLNLRGPIDA-------------------------LSSLTHL 90
+ GVTC N RV L + + L G + L+SLT L
Sbjct: 302 FSGVTCDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSL 481
Query: 91 RLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLA 150
++L++ +N +G ++ T L+ L N FSG +P EI L L L L+ N +
Sbjct: 482 KVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFS 661
Query: 151 GDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNM-TNNEFYGKVPNTMLNKF 209
G IP S +L L L N+L+G +P+ + + L EL++ +N + G +P
Sbjct: 662 GTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIP------- 820
Query: 210 GDESFSGNEGLCGSKPFQVCSLTENSPPS 238
+F E L C+LT PPS
Sbjct: 821 --PAFGSMENL-RLLEMANCNLTGEIPPS 898
Score = 74.3 bits (181), Expect = 6e-14
Identities = 50/144 (34%), Positives = 74/144 (50%), Gaps = 8/144 (5%)
Frame = +2
Query: 82 DALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLA-GNDFSGQIPPEISSLNNLL 140
++ S L L L+ N L G V SL + LK L+L N + G IPP S+ NL
Sbjct: 674 ESYSEFQSLEFLGLNANSLTGRVPESL-AKLKTLKELHLGYSNAYEGGIPPAFGSMENLR 850
Query: 141 RLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGK 200
L++++ NL G+IP + LT L +L +Q N L+G IP S M +L L+++ N+ G+
Sbjct: 851 LLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGE 1030
Query: 201 VP-------NTMLNKFGDESFSGN 217
+P N L F F G+
Sbjct: 1031IPESFSKLKNLTLMNFFQNKFRGS 1102
Score = 57.0 bits (136), Expect = 1e-08
Identities = 40/155 (25%), Positives = 71/155 (45%), Gaps = 25/155 (16%)
Frame = +2
Query: 76 NLRGPI-DALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFS-------- 126
+L G I ++ S L +L L++ N+ G++ S + + NL+ L + N+FS
Sbjct: 1016 DLTGEIPESFSKLKNLTLMNFFQNKFRGSLP-SFIGDLPNLETLQVWENNFSFVLPHNLG 1192
Query: 127 ----------------GQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQN 170
G IPP++ L ++DN G IP I +L +R+ N
Sbjct: 1193 GNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVAN 1372
Query: 171 NALSGNIPDLSSIMPNLTELNMTNNEFYGKVPNTM 205
N L G +P +P++T ++NN G++P+ +
Sbjct: 1373 NFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVI 1477
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 194 bits (492), Expect = 5e-50
Identities = 157/542 (28%), Positives = 262/542 (47%), Gaps = 13/542 (2%)
Frame = +3
Query: 139 LLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPD--LSSIMPNLTELNMTNNE 196
+ +LDLS +NL G IP+ I+ +TNL TL + +N+ G++P LSS+ L ++++ N+
Sbjct: 1599 ITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPLSSL---LISVDLSYND 1769
Query: 197 FYGKVPNTMLNKFGDES--FSGNEGLCGSKPFQVCSLTENSPPSSEPVQTVPSNPSSFPA 254
GK+P +++ +S F NE + SP P+N +S
Sbjct: 1770 LMGKLPESIVKLPHLKSLYFGCNEHM--------------SPED-------PANMNSSLI 1886
Query: 255 TSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCARGRGVNSNSLMGSEAG 314
+ R + + + G ++ I C +LL+ +F V C + L+ E
Sbjct: 1887 NTDYGRCKGKESRF---GQVIVIGAITCGSLLITLAFGVLFVCRYRQ-----KLIPWEGF 2042
Query: 315 KRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRNGFELEDLLRASAEMLGKGSL 374
K Y E + S D + L + +E ++G+G
Sbjct: 2043 AGKKYPMETNIIFSLPSKDDFFIKSVSIQAFTLEY--------IEVATERYKTLIGEGGF 2198
Query: 375 GTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLL 434
G+VYR L+DG VAVK + EF+ ++++ ++H N+V L Y ++++L
Sbjct: 2199 GSVYRGTLNDGQEVAVKVRSSTSTQGTREFDNELNLLSAIQHENLVPLLGYCNESDQQIL 2378
Query: 435 VYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKS 494
VY ++SNGSL L+G +I LDW TR+S+ LGAARGLA +HT + V H ++KS
Sbjct: 2379 VYPFMSNGSLQDRLYGEPAKRKI-LDWPTRLSIALGAARGLAYLHT-FPGRSVIHRDIKS 2552
Query: 495 SNVLLDKNGVACISDFGL---------SLLLNPVHATARLGGYRAPEQTEQKRLSQQADV 545
SN+LLD + A ++DFG S + V TA GY PE + ++LS+++DV
Sbjct: 2553 SNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGTA---GYLDPEYYKTQQLSEKSDV 2723
Query: 546 YSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQEL 605
+SFGV+LLE+++G+ P ++ T L +W +R E+ D
Sbjct: 2724 FSFGVVLLEIVSGR-----------EPLNIKRPRTEWSLVEWATPYIRGSKVDEIVDPG- 2867
Query: 606 LRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSPLCEDYDESRNSLS 665
++ E + ++ V L C+ RP+MV +V+ +ED + ++ E Y +S +SL
Sbjct: 2868 IKGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIVRELEDALIIENNASE-YMKSIDSLG 3044
Query: 666 PS 667
S
Sbjct: 3045 GS 3050
Score = 52.0 bits (123), Expect = 3e-07
Identities = 36/125 (28%), Positives = 60/125 (47%), Gaps = 1/125 (0%)
Frame = +3
Query: 43 LTNWTGPEACSASWHGVTCTPNNRVTTLVLPSLNLRGPIDALSSLTHLRLLDLHNNRLNG 102
L +W+G W G+ C +N + + LDL ++ L G
Sbjct: 1518 LESWSGDPCILLPWKGIACDGSNGSSVIT--------------------KLDLSSSNLKG 1637
Query: 103 TVSASLLSNCTNLKLLYLAGNDFSGQIPP-EISSLNNLLRLDLSDNNLAGDIPNEISRLT 161
+ +S ++ TNL+ L ++ N F G +P +SSL L+ +DLS N+L G +P I +L
Sbjct: 1638 LIPSS-IAEMTNLETLNISHNSFDGSVPSFPLSSL--LISVDLSYNDLMGKLPESIVKLP 1808
Query: 162 NLLTL 166
+L +L
Sbjct: 1809 HLKSL 1823
>TC14959 similar to GB|AAN64529.1|24797034|BT001138 At5g58299/At5g58299
{Arabidopsis thaliana;}, partial (16%)
Length = 1015
Score = 155 bits (391), Expect = 3e-38
Identities = 81/156 (51%), Positives = 112/156 (70%), Gaps = 1/156 (0%)
Frame = +2
Query: 492 VKSSNVLL-DKNGVACISDFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQQADVYSFGV 550
+KSSN+LL + A +SDFGL+ L ++ R+ GYRAPE E ++++ ++DVYSFGV
Sbjct: 2 IKSSNILLRGTDHHASVSDFGLNPLFGNGASSNRVAGYRAPEVVETRKVTFKSDVYSFGV 181
Query: 551 LLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKN 610
LLLE+LTGKAP+ + E +DLP+WV+SVVREEWT EVFD EL+R++N
Sbjct: 182 LLLELLTGKAPN-----------QASLGEEGIDLPRWVQSVVREEWTAEVFDAELMRFQN 328
Query: 611 IEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDI 646
IEEE+V +L + +ACV P++RPTM DVV+MIED+
Sbjct: 329 IEEEMVQLLQIAMACVSIVPDQRPTMQDVVRMIEDM 436
>AV419713
Length = 413
Score = 154 bits (390), Expect = 3e-38
Identities = 72/138 (52%), Positives = 105/138 (75%), Gaps = 1/138 (0%)
Frame = +3
Query: 421 KLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHT 480
+L+AYYY+K+EKL+VYDY S GS+ ++LHG RG R+PL+W TR+ + LGAARG+ARIH
Sbjct: 3 ELKAYYYSKDEKLMVYDYYSQGSVSSMLHGKRGEERVPLNWDTRLKIALGAARGIARIHV 182
Query: 481 EYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVH-ATARLGGYRAPEQTEQKRL 539
E ++ K+ HGN+KSSN+ ++ C+SD GL+ + + + +R GYRAPE T+ ++
Sbjct: 183 E-NSGKLVHGNIKSSNIFVNTKQYGCVSDLGLATMSSSLPLPISRAAGYRAPEVTDTRKA 359
Query: 540 SQQADVYSFGVLLLEVLT 557
+Q +DVYSFGV+LLE+LT
Sbjct: 360 AQPSDVYSFGVVLLELLT 413
>TC11053 weakly similar to UP|Q9FHK7 (Q9FHK7) Receptor-like protein kinase,
partial (9%)
Length = 606
Score = 152 bits (384), Expect = 2e-37
Identities = 75/89 (84%), Positives = 81/89 (90%)
Frame = +1
Query: 588 VRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIR 647
VRS+V+EEWT EVFDQELLRY+NIEEELVSML VGLACVV PEKRPTM +VVKM+EDIR
Sbjct: 1 VRSLVKEEWTAEVFDQELLRYQNIEEELVSMLQVGLACVVPLPEKRPTMAEVVKMVEDIR 180
Query: 648 VEQSPLCEDYDESRNSLSPSIPTTEDGLA 676
VE SPL EDYDESRNSLSPS+PT EDGLA
Sbjct: 181 VEPSPLGEDYDESRNSLSPSVPTPEDGLA 267
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 149 bits (375), Expect = 2e-36
Identities = 101/294 (34%), Positives = 161/294 (54%), Gaps = 12/294 (4%)
Frame = +2
Query: 365 SAEMLGKGSLGTVYRAVLDDGSTVAVKRLK-DANPCARHEFEQYMDVIGKLKHPNIVKLR 423
S ++G+GS G VY A L+DG+ VAVK+L + P +EF + ++ +LK+ N V+L
Sbjct: 365 SKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPETNNEFLTQVSMVSRLKNDNFVELH 544
Query: 424 AYYYAKEEKLLVYDYLSNGSLHALLHGNRG-----PGRIPLDWTTRISLVLGAARGLARI 478
Y ++L Y++ + GSLH +LHG +G PG LDW R+ + + AARGL +
Sbjct: 545 GYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPT-LDWIQRVRIAVDAARGLEYL 721
Query: 479 HTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSL----LLNPVHATARLG--GYRAPE 532
H + A + H +++SSNVL+ ++ A I+DF LS + +H+T LG GY APE
Sbjct: 722 HEKVQPAII-HRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHSTRVLGTFGYHAPE 898
Query: 533 QTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVV 592
+L+Q++DVYSFGV+LLE+LTG+ P + + P + L W +
Sbjct: 899 YAMTGQLTQKSDVYSFGVVLLELLTGRKP-VDHTMPRGQQ----------SLVTWATPRL 1045
Query: 593 REEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDI 646
E+ + D + L+ + + + + V CV + E RP M VVK ++ +
Sbjct: 1046SEDKVKQCVDPK-LKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKALQPL 1204
>AV416414
Length = 416
Score = 144 bits (362), Expect = 6e-35
Identities = 72/135 (53%), Positives = 98/135 (72%), Gaps = 1/135 (0%)
Frame = +2
Query: 384 DGSTVAVKRLKDANPCARHEFEQYMDVIGKL-KHPNIVKLRAYYYAKEEKLLVYDYLSNG 442
+ TV VKRLK+ + +FEQ M++ G++ +H N+V LRAYYY+K+EKLLVYDY+ G
Sbjct: 2 ESMTVVVKRLKEV-VVGKKDFEQQMEITGRVGQHANVVPLRAYYYSKDEKLLVYDYVPAG 178
Query: 443 SLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKN 502
+L +LLHG+R GR PLDW +R+ + LG ARG+A IH+ K HGN+KSSNVLL ++
Sbjct: 179 NLSSLLHGSRTGGRTPLDWDSRVKISLGTARGIAHIHS-VGGPKFTHGNIKSSNVLLSQD 355
Query: 503 GVACISDFGLSLLLN 517
CISDFGL+ L+N
Sbjct: 356 NDGCISDFGLTPLMN 400
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 142 bits (358), Expect = 2e-34
Identities = 95/285 (33%), Positives = 151/285 (52%), Gaps = 6/285 (2%)
Frame = +3
Query: 366 AEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAY 425
A +LG G G VY +D G+ VA+KR + HEF+ ++++ KL+H ++V L Y
Sbjct: 6 ALLLGVGGFGKVYYGEVDGGTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGY 185
Query: 426 YYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAA 485
E +LVYD+++ G+L L+ + P PL W R+ + +GAARGL +HT +
Sbjct: 186 CEENTEMILVYDHMAYGTLREHLYKTQKP---PLPWKQRLEICIGAARGLHYLHTG-AKY 353
Query: 486 KVPHGNVKSSNVLLDKNGVACISDFGLSL---LLNPVHATARLG---GYRAPEQTEQKRL 539
+ H +VK++N+LLD+ VA +SDFGLS L+ H + + GY PE +++L
Sbjct: 354 TIIHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQL 533
Query: 540 SQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGE 599
+ ++DVYSFGV+L E+L + PA P +E+ V L +W + +
Sbjct: 534 TDKSDVYSFGVVLFEILCAR--------PALNPSLAKEQ---VSLAEWASHCYNKGILDQ 680
Query: 600 VFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIE 644
+ D L+ K E + CV Q +RP+M DV+ +E
Sbjct: 681 ILD-PYLKGKIAPECFKKFAETAMKCVSDQGIERPSMGDVLWNLE 812
>TC10830 weakly similar to UP|Q94JL9 (Q94JL9) At1g68400/T2E12_5, partial
(24%)
Length = 702
Score = 142 bits (357), Expect = 2e-34
Identities = 83/194 (42%), Positives = 118/194 (60%), Gaps = 3/194 (1%)
Frame = +2
Query: 5 IMFFFFLFLSIYIVPCL---THNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTC 61
+ F FLS+ + P T+ D L F+ +DT +L T + C+ WHGVTC
Sbjct: 137 LFIFSLTFLSLLLTPQASTPTNPDFHPLISFKATSDTSNKLTTWNSTTHLCT--WHGVTC 310
Query: 62 TPNNRVTTLVLPSLNLRGPIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLA 121
NRV+ LVL +L+L G + L+SLT LR+L L NNR +G + LSN T ++LL+L+
Sbjct: 311 L-RNRVSRLVLENLDLHGSLHPLTSLTQLRVLSLKNNRFHGPIPN--LSNLTGMRLLFLS 481
Query: 122 GNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLS 181
N+FSG P ++SL L RLDLS N+L+G+IP ++ T LLTLRL N L G IP+++
Sbjct: 482 HNNFSGDFPVTLTSLTRLYRLDLSHNSLSGEIPAAVNNFTRLLTLRLDGNQLHGRIPNMN 661
Query: 182 SIMPNLTELNMTNN 195
PNL + N++ N
Sbjct: 662 --FPNLQDFNVSGN 697
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 141 bits (356), Expect = 3e-34
Identities = 113/361 (31%), Positives = 177/361 (48%), Gaps = 9/361 (2%)
Frame = +1
Query: 301 RGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRNGFELED 360
R +S+ G A + G K+ G G SS+G+S + F R EL D
Sbjct: 271 RSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSSNGSSNGKTAAASFGFR----ELAD 438
Query: 361 LLR--ASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPN 418
R A ++G+G G VY+ L G VAVK+L EF + ++ L H N
Sbjct: 439 ATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLSLLHHTN 618
Query: 419 IVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARI 478
+V+L Y +++LLVY+Y+ GSL L + PL+W+TR+ + +GAARGL +
Sbjct: 619 LVRLIGYCTDGDQRLLVYEYMPMGSLEDHLF-ELSHDKEPLNWSTRMKVAVGAARGLEYL 795
Query: 479 HTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPV----HATARL---GGYRAP 531
H + V + ++KS+N+LLD +SDFGL+ L PV H + R+ GY AP
Sbjct: 796 HCT-ADPPVIYRDLKSANILLDNEFNPKLSDFGLA-KLGPVGDNTHVSTRVMGTYGYCAP 969
Query: 532 EQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSV 591
E +L+ ++D+YSFGV+LLE+LTG+ + R+ E+ +L W R
Sbjct: 970 EYAMSGKLTLKSDIYSFGVVLLELLTGR-------RAIDTSRRPGEQ----NLVSWARPY 1116
Query: 592 VREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQS 651
+ LL+ + L + + C+ +QP+ RP + D+V +E + + S
Sbjct: 1117FSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLASQGS 1296
Query: 652 P 652
P
Sbjct: 1297P 1299
>BP055837
Length = 528
Score = 141 bits (355), Expect = 4e-34
Identities = 75/147 (51%), Positives = 100/147 (68%)
Frame = -1
Query: 522 TARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETV 581
T R GYRAPE ++ ++ S ++DVYSFGVLL+E+LTGK PS ++
Sbjct: 528 TGRFNGYRAPEASDGRKQSWKSDVYSFGVLLMEMLTGKCPSA-----------LDGGGEA 382
Query: 582 VDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVK 641
VDLP+WV+SVVREEWT EVFD EL+RYK+IEEE+V++L + +AC P++RP M VVK
Sbjct: 381 VDLPRWVQSVVREEWTAEVFDLELMRYKDIEEEMVALLQIAVACTAAVPDQRPRMSHVVK 202
Query: 642 MIEDIRVEQSPLCEDYDESRNSLSPSI 668
MIE++R Q C D +S S SPS+
Sbjct: 201 MIEELRGVQVSPCHDTVDS-VSESPSL 124
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 140 bits (354), Expect = 5e-34
Identities = 78/208 (37%), Positives = 124/208 (59%), Gaps = 5/208 (2%)
Frame = +2
Query: 369 LGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYA 428
LG+G G+VY DG +AVK+LK N A EF ++V+G+++H N++ LR Y
Sbjct: 320 LGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEVLGRVRHKNLLGLRGYCVG 499
Query: 429 KEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVP 488
+++L+VYDY+ N SL + LHG + L+W R+ + +G+A G+ +H E + +
Sbjct: 500 DDQRLIVYDYMPNLSLLSHLHGQFAV-EVQLNWQKRMKIAIGSAEGILYLHHEVT-PHII 673
Query: 489 HGNVKSSNVLLDKNGVACISDFGLSLLL--NPVHATARLG---GYRAPEQTEQKRLSQQA 543
H ++K+SNVLL+ + ++DFG + L+ H T R+ GY APE ++S+
Sbjct: 674 HRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSHMTTRVKGTLGYLAPEYAMWGKVSESC 853
Query: 544 DVYSFGVLLLEVLTGKAPSLQYPSPANR 571
DVYSFG+LLLE++TG+ P + P R
Sbjct: 854 DVYSFGILLLELVTGRKPIEKLPGGVKR 937
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 138 bits (348), Expect = 3e-33
Identities = 79/208 (37%), Positives = 124/208 (58%), Gaps = 5/208 (2%)
Frame = +1
Query: 369 LGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYA 428
LG+G G+VY L DGS +AVKRLK + A EF ++++ +++H N++ LR Y
Sbjct: 82 LGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEVEILARVRHKNLLSLRGYCAE 261
Query: 429 KEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVP 488
+E+L+VYDY+ N SL + LHG LDW R+++ +G+A G+ +H + + +
Sbjct: 262 GQERLIVYDYMPNLSLLSHLHGQHS-SECLLDWNRRMNIAIGSAEGIVYLHHQ-ATPHII 435
Query: 489 HGNVKSSNVLLDKNGVACISDFGLSLLL--NPVHATARLG---GYRAPEQTEQKRLSQQA 543
H ++K+SNVLLD + A ++DFG + L+ H T R+ GY APE + ++
Sbjct: 436 HRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVTTRVKGTLGYLAPEYAMLGKANECC 615
Query: 544 DVYSFGVLLLEVLTGKAPSLQYPSPANR 571
DV+SFG+LLLE+ +GK P + S R
Sbjct: 616 DVFSFGILLLELASGKKPLEKLSSTVKR 699
>TC10038 similar to UP|Q9FHK7 (Q9FHK7) Receptor-like protein kinase, partial
(16%)
Length = 601
Score = 136 bits (343), Expect = 1e-32
Identities = 69/119 (57%), Positives = 89/119 (73%)
Frame = +2
Query: 529 RAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWV 588
RA E T+ ++++Q++DVYSFGVLLLE+LTGK P L+YP E VVDLP+WV
Sbjct: 2 RATEVTDSRKITQKSDVYSFGVLLLEMLTGKTP-LRYPG----------YEDVVDLPRWV 148
Query: 589 RSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIR 647
RSVVREEWT EVFD+ELLR + +EEE+V ML + LACV + P+ RP M DVV+M+E I+
Sbjct: 149 RSVVREEWTAEVFDEELLRGQYVEEEMVQMLQIALACVAKTPDMRPRMEDVVRMVEQIK 325
>CN825263
Length = 663
Score = 127 bits (318), Expect = 8e-30
Identities = 81/194 (41%), Positives = 116/194 (59%), Gaps = 7/194 (3%)
Frame = +1
Query: 375 GTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLL 434
G VY+ +L+DG VAVK LK + EF ++++ +L H N+VKL K+ + L
Sbjct: 7 GLVYKGILNDGRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQTRCL 186
Query: 435 VYDYLSNGSLHALLHG-NRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVK 493
+Y+ + NGS+ + LHG ++ G PLDW R+ + LGAARGLA +H E S V H + K
Sbjct: 187 IYELVPNGSVESHLHGADKETG--PLDWNARMKIALGAARGLAYLH-EDSNPCVIHRDFK 357
Query: 494 SSNVLLDKNGVACISDFGLSLLL----NPVHATARLG--GYRAPEQTEQKRLSQQADVYS 547
SSN+LL+ + +SDFGL+ N +T +G GY APE L ++DVYS
Sbjct: 358 SSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYS 537
Query: 548 FGVLLLEVLTGKAP 561
+GV+LLE+LTG P
Sbjct: 538 YGVVLLELLTGTKP 579
>TC16505 similar to GB|AAN64529.1|24797034|BT001138 At5g58299/At5g58299
{Arabidopsis thaliana;}, partial (16%)
Length = 649
Score = 126 bits (317), Expect = 1e-29
Identities = 64/120 (53%), Positives = 86/120 (71%)
Frame = +2
Query: 543 ADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFD 602
+DVYSFGVLLLE+LTGKAP PSP + +VDLP WV+SVVREEWT EVFD
Sbjct: 2 SDVYSFGVLLLEMLTGKAP---LPSPGR--------DDMVDLPSWVQSVVREEWTAEVFD 148
Query: 603 QELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSPLCEDYDESRN 662
EL++Y+N+EEE+V ML + +ACV + P+ RP+M +VV+MIE+IR S +E+++
Sbjct: 149 VELMKYQNLEEEMVQMLQIAMACVAKMPDMRPSMDEVVRMIEEIRPSDSENRPSSEENKS 328
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 125 bits (313), Expect = 3e-29
Identities = 91/281 (32%), Positives = 144/281 (50%), Gaps = 6/281 (2%)
Frame = +1
Query: 369 LGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYA 428
LG+G G VY+ +L +G +AVKRL + + EF+ + +I +L+H N+VKL
Sbjct: 1690 LGEGGFGPVYKGLLANGQEIAVKRLSNTSGQGMEEFKNEIKLIARLQHRNLVKLFGCSVH 1869
Query: 429 KEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVP 488
++E + +N + LL R +DW R+ ++ G ARGL +H + S ++
Sbjct: 1870 QDE-----NSHANKKMKILLDSTRSK---LVDWNKRLQIIDGIARGLLYLHQD-SRLRII 2022
Query: 489 HGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLG------GYRAPEQTEQKRLSQQ 542
H ++K+SN+LLD ISDFGL+ + AR GY PE S +
Sbjct: 2023 HRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMGTYGYMPPEYAVHGSFSIK 2202
Query: 543 ADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFD 602
+DV+SFGV++LE+++GK Y P + + + W+ EE E+ D
Sbjct: 2203 SDVFSFGVIVLEIISGKKIGRFY-DPHHHLNLLSHAWRL-----WI-----EERPLELVD 2349
Query: 603 QELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMI 643
ELL I E++ +HV L CV ++PE RP M+ +V M+
Sbjct: 2350 -ELLDDPVIPTEILRYIHVALLCVQRRPENRPDMLSIVLML 2469
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,547,489
Number of Sequences: 28460
Number of extensions: 164285
Number of successful extensions: 1879
Number of sequences better than 10.0: 399
Number of HSP's better than 10.0 without gapping: 1512
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1608
length of query: 676
length of database: 4,897,600
effective HSP length: 96
effective length of query: 580
effective length of database: 2,165,440
effective search space: 1255955200
effective search space used: 1255955200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC139525.8


