

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139525.4 + phase: 0
(490 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
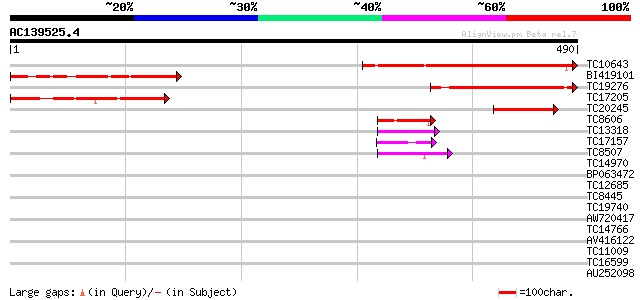
Score E
Sequences producing significant alignments: (bits) Value
TC10643 similar to UP|BAC84847 (BAC84847) Acidic nuclear phospho... 250 4e-67
BI419101 174 4e-44
TC19276 160 3e-40
TC17205 weakly similar to PIR|S50830|S50830 Machado-Joseph disea... 126 7e-30
TC20245 70 8e-13
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 44 5e-05
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 42 2e-04
TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly pr... 42 2e-04
TC8507 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetyla... 40 7e-04
TC14970 similar to UP|O24294 (O24294) Legumin (Minor small) prec... 38 0.004
BP063472 37 0.007
TC12685 weakly similar to UP|Q7QA92 (Q7QA92) AgCP13961 (Fragment... 37 0.007
TC8445 similar to UP|O80910 (O80910) At2g38410 protein, partial ... 37 0.007
TC19740 similar to UP|FKH_DROME (P14734) Fork head protein, part... 37 0.010
AW720417 37 0.010
TC14766 similar to UP|Q7XXR8 (Q7XXR8) Nascent polypeptide associ... 36 0.012
AV416122 35 0.021
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 35 0.021
TC16599 similar to UP|Q8W239 (Q8W239) GT-2 factor (Fragment), pa... 35 0.028
AU252098 35 0.036
>TC10643 similar to UP|BAC84847 (BAC84847) Acidic nuclear
phosphoprotein-like protein, partial (5%)
Length = 789
Score = 250 bits (638), Expect = 4e-67
Identities = 128/188 (68%), Positives = 160/188 (85%), Gaps = 3/188 (1%)
Frame = +2
Query: 306 EGLRMLKIGSGYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRK 365
EGLRM+K+GSG EE++DE+EE++SE+ SD+ EDES EG S+G + D+DE +N G+ SRK
Sbjct: 5 EGLRMMKLGSG--EEDEDEEEEEDSEESSDDDEDESAEGGSRGTLVDEDENDN-GRSSRK 175
Query: 366 RARKG-GFSFPRSSSSTQLVNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYE 424
R RK G S SS+ +QLV Q+N E+SGV QDGGKS WEK+QW+R+RIMQLEEQ++ Y+
Sbjct: 176 RTRKSAGCSVSVSSTPSQLVQQLNTEVSGVLQDGGKSPWEKRQWMRSRIMQLEEQQLSYQ 355
Query: 425 SQAFQLEKQRLKWARYSSKKEREMERAKLENERRRLENERMVLLIRKKELELMHI--QQQ 482
+QAF LEKQR+KWAR+SSKKEREMERAKLENE+RRLENERMVL+I +KELELM++ QQQ
Sbjct: 356 TQAFDLEKQRVKWARFSSKKEREMERAKLENEKRRLENERMVLIIHQKELELMNLQHQQQ 535
Query: 483 QQQQHSST 490
QQQQHSST
Sbjct: 536 QQQQHSST 559
>BI419101
Length = 560
Score = 174 bits (440), Expect = 4e-44
Identities = 96/148 (64%), Positives = 107/148 (71%)
Frame = +2
Query: 1 MFSTITPSGLLGVVDNIPLHQQQQQQKQQNLPNQQQQNPHQLHHSQVVSYAPQHHDTDTN 60
MFST SGLLGV +N +QQNL NQQ NPH LHH Q+VSY D
Sbjct: 164 MFSTSISSGLLGVENN------NNPLQQQNLTNQQ--NPHHLHHPQMVSY-------DQQ 298
Query: 61 QHHHHSMKHGFPPFSSSKNKQQQQSSQMSDEDEPNFPAEESSGGDPKRKISPWQRMKWTD 120
Q +KHG+ P+SS+KNKQQQ S +SD+DE FPA+E+SG DPKRKISPWQRMKWTD
Sbjct: 299 QQLQQPIKHGYSPYSSAKNKQQQSS--ISDDDELGFPADETSG-DPKRKISPWQRMKWTD 469
Query: 121 TMVRLLIMAVYYIGDEAGSEGTDPNKKK 148
TMVRLLIMAVYYIGDEAGSEGTDPN KK
Sbjct: 470 TMVRLLIMAVYYIGDEAGSEGTDPNGKK 553
>TC19276
Length = 494
Score = 160 bits (406), Expect = 3e-40
Identities = 80/127 (62%), Positives = 104/127 (80%)
Frame = +2
Query: 364 RKRARKGGFSFPRSSSSTQLVNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGY 423
RKRARK G +Q++ Q+++E+SGV QDGGKS WEKKQW++ R++QL+EQ++ Y
Sbjct: 23 RKRARKSGVM------PSQVMXQLSSEVSGVLQDGGKSAWEKKQWMKKRMVQLDEQQVSY 184
Query: 424 ESQAFQLEKQRLKWARYSSKKEREMERAKLENERRRLENERMVLLIRKKELELMHIQQQQ 483
++AF+LEK RLKWAR+SS+KEREMER +LENER RLE ERM+LL+R +ELELM+I QQ
Sbjct: 185 RAEAFELEKPRLKWARFSSQKEREMERHQLENERSRLEIERMILLLRHQELELMNI--QQ 358
Query: 484 QQQHSST 490
QQQHSST
Sbjct: 359 QQQHSST 379
>TC17205 weakly similar to PIR|S50830|S50830 Machado-Joseph disease MJD1a
protein - human {Homo sapiens;}, partial (7%)
Length = 528
Score = 126 bits (317), Expect = 7e-30
Identities = 73/150 (48%), Positives = 92/150 (60%), Gaps = 12/150 (8%)
Frame = +1
Query: 1 MFSTITPSGLLGVVDNIPLHQQQQQQKQQNLPNQQQQNPHQLHH-SQVVSYAPQHHDTDT 59
+F TI+ SG LGV + PL+QQ Q NP LHH SQ++SYA HHD +T
Sbjct: 121 IFPTISSSGWLGVENQNPLNQQNHHQ-----------NPLPLHHHSQMLSYATTHHD-NT 264
Query: 60 NQHHHHSMKHGFP-----------PFSSSKNKQQQQSSQMSDEDEPNFPAEESSGGDPKR 108
+ H +KHG+P +++ N + Q + + DEDE F A+++S DPKR
Sbjct: 265 DTHIQQPIKHGYPYSAKTNNNNSNGTNNNSNNKAQSNINLRDEDE--FAADDNSSADPKR 438
Query: 109 KISPWQRMKWTDTMVRLLIMAVYYIGDEAG 138
K SPW RMKWTDTMVRLLIMAVYYIGDEAG
Sbjct: 439 KNSPWHRMKWTDTMVRLLIMAVYYIGDEAG 528
>TC20245
Length = 443
Score = 70.1 bits (170), Expect = 8e-13
Identities = 30/56 (53%), Positives = 45/56 (79%)
Frame = +1
Query: 419 QKIGYESQAFQLEKQRLKWARYSSKKEREMERAKLENERRRLENERMVLLIRKKEL 474
QK+ +++ +LEK RLKW R+S KK+RE+E+ KLENER +LENER+ L +++KE+
Sbjct: 4 QKLQIQAEMLELEKPRLKWQRFSKKKDRELEKMKLENERMKLENERIALELKRKEI 171
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 44.3 bits (103), Expect = 5e-05
Identities = 22/53 (41%), Positives = 34/53 (63%), Gaps = 3/53 (5%)
Frame = -2
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDG---KPSRKRAR 368
E++DD+DE+D+ ED D GEDE EG + D++E+E D +P +KR +
Sbjct: 398 EDDDDDDEDDDEED--DGGEDEEEEGVEEEDNEDEEEDEEDEEALQPPKKRKK 246
Score = 27.7 bits (60), Expect = 4.4
Identities = 14/51 (27%), Positives = 27/51 (52%)
Frame = -2
Query: 321 EDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGG 371
+D+ DE+D+ + EGE++ G+ N+ + + N K+A +GG
Sbjct: 602 DDEGDEDDDGDGPFGEGEEDLSSEDGAGYGNNSNNKSNS-----KKAPEGG 465
Score = 27.3 bits (59), Expect = 5.8
Identities = 12/35 (34%), Positives = 15/35 (42%)
Frame = +2
Query: 31 LPNQQQQNPHQLHHSQVVSYAPQHHDTDTNQHHHH 65
LP Q PH LHH + + + H HH H
Sbjct: 314 LPLQHPLPPHLLHHHPLHRHLHRRHHHPLGPHHLH 418
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 42.0 bits (97), Expect = 2e-04
Identities = 18/53 (33%), Positives = 31/53 (57%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGG 371
EEE+D+DEE++ ED D+ +D+ + + +D D+EE +G + K G
Sbjct: 252 EEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEVEVEGKDG 410
Score = 36.2 bits (82), Expect = 0.012
Identities = 15/49 (30%), Positives = 28/49 (56%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRA 367
EE+D+++++D+ +D D+GE+E + +E E +GK K A
Sbjct: 276 EEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEVEVEGKDGAKGA 422
Score = 34.7 bits (78), Expect = 0.036
Identities = 17/65 (26%), Positives = 32/65 (49%), Gaps = 5/65 (7%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARK-----GGFS 373
+++DD+D++D E+ D+ +DE EG + + +D + + K A G FS
Sbjct: 300 DDDDDDDDDDGEEEEDDDDDDEEEEGSEEVEVEGKDGAKGAAAQTTKAAAAMCVGIGSFS 479
Query: 374 FPRSS 378
P +
Sbjct: 480 DPNEA 494
Score = 32.3 bits (72), Expect = 0.18
Identities = 14/42 (33%), Positives = 26/42 (61%)
Frame = +3
Query: 320 EEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGK 361
+ D+E+EED+ E+ DE ED+ +D D++++DG+
Sbjct: 240 QTDNEEEEDDDEEEDDEDEDD----------DDDDDDDDDGE 335
Score = 27.7 bits (60), Expect = 4.4
Identities = 13/26 (50%), Positives = 19/26 (73%), Gaps = 2/26 (7%)
Frame = +3
Query: 316 GYVEEEDDEDEEDE--SEDFSDEGED 339
G EE+DD+D+E+E SE+ EG+D
Sbjct: 330 GEEEEDDDDDDEEEEGSEEVEVEGKD 407
Score = 27.7 bits (60), Expect = 4.4
Identities = 12/34 (35%), Positives = 15/34 (43%)
Frame = -1
Query: 39 PHQLHHSQVVSYAPQHHDTDTNQHHHHSMKHGFP 72
PH HH ++ HH + HHHH H P
Sbjct: 366 PHHHHHHLLLPPHHHHH----HHHHHHLHLHHLP 277
>TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly protein
1-like protein 3, partial (81%)
Length = 1298
Score = 42.0 bits (97), Expect = 2e-04
Identities = 19/52 (36%), Positives = 31/52 (59%)
Frame = +2
Query: 318 VEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARK 369
+EE+D++ +EDE ED D+ E+E + +D+D+EE +GK K K
Sbjct: 770 IEEDDEDGDEDEDEDDDDDEEEEDDDD------DDEDDEEGEGKSKSKSGSK 907
Score = 38.9 bits (89), Expect = 0.002
Identities = 19/68 (27%), Positives = 36/68 (52%)
Frame = +2
Query: 315 SGYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSF 374
+G E+ D ED E++ ED DE EDE + + +D D+E+++ + +++ G +
Sbjct: 737 TGEAEQSDFEDIEEDDED-GDEDEDEDDDDDEEEEDDDDDDEDDEEGEGKSKSKSGSKAR 913
Query: 375 PRSSSSTQ 382
P T+
Sbjct: 914 PGKDQPTE 937
>TC8507 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (40%)
Length = 620
Score = 40.4 bits (93), Expect = 7e-04
Identities = 21/66 (31%), Positives = 32/66 (47%), Gaps = 2/66 (3%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEE--NDGKPSRKRARKGGFSFPR 376
+EEDD D+ED+ +D SDE D++ + +D DEE +KRA P
Sbjct: 3 DEEDDSDDEDDEDDDSDEEMDDADSDSDESDSDDTDEETPVKKADQGKKRANDSALKTPV 182
Query: 377 SSSSTQ 382
S+ +
Sbjct: 183 STKKAK 200
Score = 28.9 bits (63), Expect = 2.0
Identities = 12/34 (35%), Positives = 16/34 (46%)
Frame = -3
Query: 36 QQNPHQLHHSQVVSYAPQHHDTDTNQHHHHSMKH 69
Q + HQ HH + QHH + HH H + H
Sbjct: 123 QGSLHQYHHCRSHHCHYQHHPSPHQNHHLHHLHH 22
>TC14970 similar to UP|O24294 (O24294) Legumin (Minor small) precursor,
partial (51%)
Length = 1566
Score = 37.7 bits (86), Expect = 0.004
Identities = 25/111 (22%), Positives = 45/111 (40%)
Frame = +1
Query: 258 QVEGGSGTTTPPQNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSGY 317
+VEG + P++ + + + + +QH HS +G + SR
Sbjct: 460 KVEGDDLSFISPESADEDEDEEDEDEDQEQHGRHSRPSGRRTRRPSR------------- 600
Query: 318 VEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRAR 368
E++EDE ED + G G + H ++EEE + + R R +
Sbjct: 601 ------EEDEDEDEDEEERQGGRRGPGERQRHFEREEEEEREERGPRGRGK 735
Score = 37.0 bits (84), Expect = 0.007
Identities = 32/125 (25%), Positives = 52/125 (41%), Gaps = 1/125 (0%)
Frame = +1
Query: 247 HGGVATSNVQHQVEGGSGTTTPPQNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGE 306
HG + Q + EGGS + QQ H Q S ++ + G+
Sbjct: 304 HGQHHQESEQEEEEGGSILSGFGAEFLQQVFNIDHDTAKQ---LQSPDDQRRQIVKVEGD 474
Query: 307 GLRMLKIGSGYVEEEDDEDEEDESEDFSDEGEDESGEG-CSKGHINDQDEEENDGKPSRK 365
L + S ++DEDEEDE ED G G ++ ++DE+E++ + R+
Sbjct: 475 DLSFISPESA----DEDEDEEDEDEDQEQHGRHSRPSGRRTRRPSREEDEDEDEDEEERQ 642
Query: 366 RARKG 370
R+G
Sbjct: 643 GGRRG 657
>BP063472
Length = 459
Score = 37.0 bits (84), Expect = 0.007
Identities = 23/70 (32%), Positives = 34/70 (47%), Gaps = 6/70 (8%)
Frame = -3
Query: 17 IPLHQQQQQQKQQNLPNQQQQNP-----HQLHHSQVVSYAPQHHDTD-TNQHHHHSMKHG 70
+P H Q +Q+ P + +QNP H Q++ PQ H + T+QHHH +H
Sbjct: 259 LPKHPQSKQKLLLLFPQKHKQNPISQPNHPQSKQQLLLLFPQKHKQNPTSQHHHQKAQHF 80
Query: 71 FPPFSSSKNK 80
SS+K K
Sbjct: 79 LA--SSTKRK 56
>TC12685 weakly similar to UP|Q7QA92 (Q7QA92) AgCP13961 (Fragment), partial
(9%)
Length = 547
Score = 37.0 bits (84), Expect = 0.007
Identities = 44/163 (26%), Positives = 69/163 (41%), Gaps = 20/163 (12%)
Frame = +2
Query: 268 PPQNQP---QQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSGYVEEEDDE 324
PP++ P Q H+H+ N+ FH R R V+EEDDE
Sbjct: 59 PPKSFPLSCQNPSSHRHKHPNRAAPFHH-----------RPPTTRYK------VDEEDDE 187
Query: 325 DEED--ESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFS--------- 373
+ ED E +D++D +D++ E G+ EE+DG +KR +GG S
Sbjct: 188 EFEDAKEFDDYND--DDDAPENGYAGNF-----EEDDGFARKKRKLRGGPSNYEFAPRAK 346
Query: 374 FP------RSSSSTQLVNQMNNEISGVFQDGGKSTWEKKQWIR 410
FP SS++ + + V+ D S WE++ +R
Sbjct: 347 FPFGNRGGSSSAAEEWTEHATLVLLEVWGDKVSSNWEEQFEVR 475
>TC8445 similar to UP|O80910 (O80910) At2g38410 protein, partial (7%)
Length = 841
Score = 37.0 bits (84), Expect = 0.007
Identities = 20/55 (36%), Positives = 24/55 (43%), Gaps = 1/55 (1%)
Frame = +1
Query: 41 QLHHS-QVVSYAPQHHDTDTNQHHHHSMKHGFPPFSSSKNKQQQQSSQMSDEDEP 94
QLH+ Q + Y PQ H QHH H H P S Q Q SQ + +P
Sbjct: 10 QLHNEPQPLQYQPQPHLQHQPQHHPHFQPHPQPQLQSQPQPQHIQQSQPQPQSQP 174
>TC19740 similar to UP|FKH_DROME (P14734) Fork head protein, partial (4%)
Length = 622
Score = 36.6 bits (83), Expect = 0.010
Identities = 34/129 (26%), Positives = 51/129 (39%), Gaps = 4/129 (3%)
Frame = +3
Query: 2 FSTITPSGLLGVVDNIPLHQQQQQQKQQNLPNQQQQNPHQLHHSQVVSYAPQHH---DTD 58
F TP ++ + P HQQ Q Q+ Q+ QQQQ P HH Q P+ H
Sbjct: 66 FLHFTPIAMMYNDPHNPQHQQHQHQQHQH-QFQQQQQPQHFHHQQP---GPEFHRGPPPP 233
Query: 59 TNQHHHHSMKHGFPPFSSSKNKQQQQSSQMSDEDEPNFPA-EESSGGDPKRKISPWQRMK 117
Q H P SS+ + P++ +S G RK + + +
Sbjct: 234 PPQQPHPPPMMRQPSASSTNLAPEFHHPGPGGPPPPHYDVHSDSHGAKRMRKHTQRKAVD 413
Query: 118 WTDTMVRLL 126
+T T+VR +
Sbjct: 414 YTSTVVRYM 440
Score = 33.1 bits (74), Expect = 0.11
Identities = 14/27 (51%), Positives = 15/27 (54%)
Frame = +3
Query: 270 QNQPQQQHQHQHQQQNQQHCFHSSENG 296
Q QQHQHQ QQQ Q FH + G
Sbjct: 123 QQHQHQQHQHQFQQQQQPQHFHHQQPG 203
>AW720417
Length = 554
Score = 36.6 bits (83), Expect = 0.010
Identities = 29/72 (40%), Positives = 32/72 (44%), Gaps = 4/72 (5%)
Frame = +3
Query: 20 HQQQQQQKQ----QNLPNQQQQNPHQLHHSQVVSYAPQHHDTDTNQHHHHSMKHGFPPFS 75
HQQQQQQ Q +PNQQQ HQ H Q PQ+H QH H
Sbjct: 30 HQQQQQQHQYVQRHMMPNQQQY--HQQQHQQ---NQPQYH----QQHQH----------- 149
Query: 76 SSKNKQQQQSSQ 87
N+QQQQ Q
Sbjct: 150---NQQQQQQQQ 176
Score = 33.5 bits (75), Expect = 0.081
Identities = 23/69 (33%), Positives = 31/69 (44%), Gaps = 2/69 (2%)
Frame = +3
Query: 270 QNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSG--YVEEEDDEDEE 327
QNQPQ QHQH QQ QQ + + LR ++G V EE ++ +
Sbjct: 114 QNQPQYHQQHQHNQQQQQQ--------------QQQQWLRRTQLGGNDTNVVEEVEKTVQ 251
Query: 328 DESEDFSDE 336
ES D S +
Sbjct: 252 SESADPSSQ 278
Score = 31.6 bits (70), Expect = 0.31
Identities = 17/63 (26%), Positives = 24/63 (37%)
Frame = +3
Query: 29 QNLPNQQQQNPHQLHHSQVVSYAPQHHDTDTNQHHHHSMKHGFPPFSSSKNKQQQQSSQM 88
Q P+QQQQ HQ Y +H + Q+H + P + QQQ Q
Sbjct: 18 QPRPHQQQQQQHQ--------YVQRHMMPNQQQYHQQQHQQNQPQYHQQHQHNQQQQQQQ 173
Query: 89 SDE 91
+
Sbjct: 174QQQ 182
>TC14766 similar to UP|Q7XXR8 (Q7XXR8) Nascent polypeptide associated
complex alpha chain, partial (58%)
Length = 600
Score = 36.2 bits (82), Expect = 0.012
Identities = 24/72 (33%), Positives = 35/72 (48%)
Frame = +2
Query: 318 VEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFPRS 377
V E+DDED++DE +D D+ E EG + G + ++ K SRK K G P +
Sbjct: 131 VVEDDDEDDDDEDDDDEDDDNIEGQEGDASG----RSKQTRSEKKSRKAMLKLGMK-PVT 295
Query: 378 SSSTQLVNQMNN 389
S V + N
Sbjct: 296 GVSRVTVKKSKN 331
>AV416122
Length = 414
Score = 35.4 bits (80), Expect = 0.021
Identities = 14/28 (50%), Positives = 21/28 (75%)
Frame = +3
Query: 316 GYVEEEDDEDEEDESEDFSDEGEDESGE 343
G EE+D+ED+E+E E+ DE +D +GE
Sbjct: 300 GEEEEDDEEDKEEEDEEEDDEDDDSNGE 383
Score = 30.8 bits (68), Expect = 0.52
Identities = 11/26 (42%), Positives = 18/26 (68%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEG 344
EEE+D++E+ E ED ++ ED+ G
Sbjct: 303 EEEEDDEEDKEEEDEEEDDEDDDSNG 380
Score = 30.4 bits (67), Expect = 0.68
Identities = 13/27 (48%), Positives = 20/27 (73%), Gaps = 1/27 (3%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGE-DESGEG 344
+EE+DE+E+DE +D + E E D G+G
Sbjct: 330 KEEEDEEEDDEDDDSNGEAEVDRKGKG 410
Score = 28.5 bits (62), Expect = 2.6
Identities = 15/52 (28%), Positives = 26/52 (49%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKG 370
+ E D D + E E+ D+ ED+ E ++D+E++D + RKG
Sbjct: 267 DNEPDNDVQFEGEEEEDDEEDKEEED------EEEDDEDDDSNGEAEVDRKG 404
Score = 27.7 bits (60), Expect = 4.4
Identities = 13/43 (30%), Positives = 24/43 (55%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGK 361
E E++ED+E++ E+ +E +DE + N + E + GK
Sbjct: 297 EGEEEEDDEEDKEEEDEEEDDEDDDS------NGEAEVDRKGK 407
Score = 27.3 bits (59), Expect = 5.8
Identities = 16/49 (32%), Positives = 26/49 (52%)
Frame = +3
Query: 322 DDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKG 370
+ E+EED+ ED +E E+E +D+D++ N G+ R KG
Sbjct: 297 EGEEEEDDEEDKEEEDEEE----------DDEDDDSN-GEAEVDRKGKG 410
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 35.4 bits (80), Expect = 0.021
Identities = 16/49 (32%), Positives = 29/49 (58%)
Frame = +2
Query: 323 DEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGG 371
DED++D+ ED D+G+++ + G +D DEE+ + + + R GG
Sbjct: 26 DEDDDDDGED--DDGDEDDDDDAPGGGDDDDDEEDEEEEGGVEGGRGGG 166
Score = 33.1 bits (74), Expect = 0.11
Identities = 16/42 (38%), Positives = 24/42 (57%)
Frame = +2
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDG 360
+E+DD+D ED D DE +D+ G +++DEEE G
Sbjct: 26 DEDDDDDGED---DDGDEDDDDDAPGGGDDDDDEEDEEEEGG 142
Score = 32.7 bits (73), Expect = 0.14
Identities = 16/58 (27%), Positives = 30/58 (51%), Gaps = 6/58 (10%)
Frame = +2
Query: 319 EEEDDE------DEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKG 370
+E+DD+ D++D+ ED +EG E G G +D D++++D + + G
Sbjct: 65 DEDDDDDAPGGGDDDDDEEDEEEEGGVEGGRGGGGDPDDDDDDDDDDDEEEEEEEDLG 238
Score = 30.8 bits (68), Expect = 0.52
Identities = 14/50 (28%), Positives = 26/50 (52%)
Frame = +2
Query: 326 EEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFP 375
+ED+ +D D+ DE + + G +D D+EE++ + +GG P
Sbjct: 26 DEDDDDDGEDDDGDEDDDDDAPGGGDDDDDEEDEEEEGGVEGGRGGGGDP 175
Score = 30.4 bits (67), Expect = 0.68
Identities = 20/55 (36%), Positives = 26/55 (46%), Gaps = 1/55 (1%)
Frame = +2
Query: 318 VEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPS-RKRARKGG 371
V +DE+ + E DEG+D ND D+ E G PS RKR+ K G
Sbjct: 263 VAAAEDEEASSDFEPEEDEGDD-----------NDNDDGEKAGVPSKRKRSDKDG 394
>TC16599 similar to UP|Q8W239 (Q8W239) GT-2 factor (Fragment), partial (37%)
Length = 693
Score = 35.0 bits (79), Expect = 0.028
Identities = 20/75 (26%), Positives = 39/75 (51%), Gaps = 14/75 (18%)
Frame = +1
Query: 134 GDEAGSEGTDPNKKKSSGLLQ--------------KKGKWKSVSRAMMEKGFYVSPQQCE 179
GD+ GS G+ ++++ LL+ K W+ VSR + G++ S ++C+
Sbjct: 172 GDKIGSGGSRWPRQETLALLKIRSDMDAVFRDSSLKGPLWEEVSRKLAGLGYHRSAKKCK 351
Query: 180 DKFNDLNKRYKRVND 194
+KF ++ K +KR +
Sbjct: 352 EKFENVYKYHKRTKE 396
>AU252098
Length = 350
Score = 34.7 bits (78), Expect = 0.036
Identities = 20/71 (28%), Positives = 30/71 (42%), Gaps = 1/71 (1%)
Frame = -1
Query: 35 QQQNPHQLHHSQVVSYAPQHHDTDTNQHHHHSMKHGFPPFSSSKNKQQQ-QSSQMSDEDE 93
Q+ H HH + P H + +N HH P SS Q Q Q + + E
Sbjct: 335 QKTKHHGPHHYRHHQENPPTHQSHSNSTHHQQEGGPHHPLPSSHQHQNQLQHQRAASEAA 156
Query: 94 PNFPAEESSGG 104
P+FP +++ G
Sbjct: 155 PHFPVSKTTTG 123
Score = 28.1 bits (61), Expect = 3.4
Identities = 15/47 (31%), Positives = 20/47 (41%)
Frame = -1
Query: 252 TSNVQHQVEGGSGTTTPPQNQPQQQHQHQHQQQNQQHCFHSSENGVG 298
+++ HQ EGG P +Q Q Q QHQ F S+ G
Sbjct: 263 SNSTHHQQEGGPHHPLPSSHQHQNQLQHQRAASEAAPHFPVSKTTTG 123
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.310 0.128 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,179,820
Number of Sequences: 28460
Number of extensions: 153334
Number of successful extensions: 3167
Number of sequences better than 10.0: 285
Number of HSP's better than 10.0 without gapping: 2113
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2709
length of query: 490
length of database: 4,897,600
effective HSP length: 94
effective length of query: 396
effective length of database: 2,222,360
effective search space: 880054560
effective search space used: 880054560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC139525.4


