

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139525.10 + phase: 0
(125 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
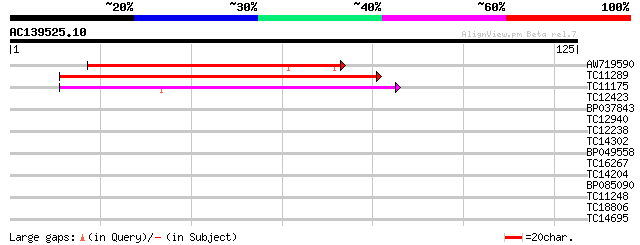
Score E
Sequences producing significant alignments: (bits) Value
AW719590 63 2e-11
TC11289 weakly similar to UP|Q9LUE5 (Q9LUE5) Emb|CAB86483.1, par... 62 2e-11
TC11175 weakly similar to GB|AAL06796.1|15809736|AY054135 AT3g60... 55 3e-09
TC12423 similar to PIR|T01558|T01558 auxin-induced protein homol... 35 0.003
BP037843 32 0.041
TC12940 31 0.054
TC12238 29 0.20
TC14302 similar to UP|Y230_ARATH (O80934) Protein At2g37660, chl... 26 1.7
BP049558 26 2.3
TC16267 similar to GB|AAP68282.1|31711852|BT008843 At1g73060 {Ar... 25 2.9
TC14204 homologue to UP|Q9AVG8 (Q9AVG8) Isopentenyl diphosphate ... 24 6.6
BP085090 24 8.6
TC11248 homologue to UP|P93323 (P93323) Cdc2MsF protein, partial... 24 8.6
TC18806 similar to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A lig... 24 8.6
TC14695 similar to UP|Q9FQE5 (Q9FQE5) Glutathione S-transferase ... 24 8.6
>AW719590
Length = 496
Score = 62.8 bits (151), Expect = 2e-11
Identities = 34/60 (56%), Positives = 42/60 (69%), Gaps = 3/60 (5%)
Frame = +2
Query: 18 DVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF-NKSGPLSIPC--DEFL 74
+VP+GHLAV VG+ RRFVI YLN P QELL QA E +G+ + +G L IPC D+FL
Sbjct: 137 EVPKGHLAVYVGDEMRRFVIPVSYLNQPSFQELLYQAEEEFGYDHPTGGLKIPCREDDFL 316
>TC11289 weakly similar to UP|Q9LUE5 (Q9LUE5) Emb|CAB86483.1, partial (32%)
Length = 765
Score = 62.4 bits (150), Expect = 2e-11
Identities = 26/71 (36%), Positives = 45/71 (62%)
Frame = +2
Query: 12 GKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCD 71
GK P G +V VG+ +RFVI+ +Y+NHP+ + LL++A YG++ GP+ +PC+
Sbjct: 281 GKSTTVVAPEGCFSVYVGQQMQRFVIKTEYVNHPLFKMLLEEAESEYGYSSQGPIVLPCN 460
Query: 72 EFLFEDILLSL 82
+F +L+ +
Sbjct: 461 VDVFYKVLMEM 493
>TC11175 weakly similar to GB|AAL06796.1|15809736|AY054135
AT3g60690/T4C21_100 {Arabidopsis thaliana;}, partial
(42%)
Length = 692
Score = 55.5 bits (132), Expect = 3e-09
Identities = 28/78 (35%), Positives = 42/78 (52%), Gaps = 3/78 (3%)
Frame = +1
Query: 12 GKKLPSDVPRGHLAVTVGETN---RRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSI 68
G L VP+GHL V VGE + R ++ Y NHP+ ELL + + +GF+ G ++I
Sbjct: 118 GSNLDPKVPKGHLPVYVGEEDGELHRLLVPVIYFNHPLFGELLKETEDEFGFHHPGGITI 297
Query: 69 PCDEFLFEDILLSLGGGT 86
PC FE + + G+
Sbjct: 298 PCRVTEFERVKTRIAAGS 351
>TC12423 similar to PIR|T01558|T01558 auxin-induced protein homolog
A_TM018A10.6 - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (43%)
Length = 317
Score = 35.4 bits (80), Expect = 0.003
Identities = 19/55 (34%), Positives = 29/55 (52%), Gaps = 2/55 (3%)
Frame = +3
Query: 18 DVPRGHLAVTVG--ETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPC 70
++P+G LA+ VG E RFVI D L L ++ ++ GP++IPC
Sbjct: 129 NIPKGCLAIMVGQGEEQERFVIPVDLHQPSTLHALAERG*RRIWVDQKGPITIPC 293
>BP037843
Length = 443
Score = 31.6 bits (70), Expect = 0.041
Identities = 14/30 (46%), Positives = 22/30 (72%)
Frame = -2
Query: 50 LLDQAYEGYGFNKSGPLSIPCDEFLFEDIL 79
LL++A E +GF+ SG L+IPC+ F+ +L
Sbjct: 436 LLEKAAEEFGFDHSGALTIPCEIETFKYLL 347
>TC12940
Length = 480
Score = 31.2 bits (69), Expect = 0.054
Identities = 20/57 (35%), Positives = 27/57 (47%)
Frame = +2
Query: 60 FNKSGPLSIPCDEFLFEDILLSLGGGTVARRSSSPVLTKKLDLSFLKDAVPLLEAFD 116
F SG S C F D LL + +V + PVL LD +LKD++ +FD
Sbjct: 98 FQDSGYCSGRCCSFWILDCLLDVSQVSVEEKKW*PVLRLNLDCIYLKDSMMHRSSFD 268
>TC12238
Length = 564
Score = 29.3 bits (64), Expect = 0.20
Identities = 12/23 (52%), Positives = 17/23 (73%)
Frame = +1
Query: 13 KKLPSDVPRGHLAVTVGETNRRF 35
+K P+ VP+GHLAV VG+ + F
Sbjct: 484 EKTPAAVPKGHLAVYVGQKDGEF 552
>TC14302 similar to UP|Y230_ARATH (O80934) Protein At2g37660, chloroplast
precursor, partial (75%)
Length = 1066
Score = 26.2 bits (56), Expect = 1.7
Identities = 17/58 (29%), Positives = 27/58 (46%)
Frame = -1
Query: 57 GYGFNKSGPLSIPCDEFLFEDILLSLGGGTVARRSSSPVLTKKLDLSFLKDAVPLLEA 114
G+GF+ + +P DEF I + G R ++ + L LSF VP+ +A
Sbjct: 655 GFGFSLEKLIILP*DEFPKTSIFI*QATGPYDRVWNTRISKVLLSLSFPNQYVPITQA 482
>BP049558
Length = 275
Score = 25.8 bits (55), Expect = 2.3
Identities = 14/31 (45%), Positives = 17/31 (54%), Gaps = 1/31 (3%)
Frame = +1
Query: 5 WRKNACSGKKLPSDVPRGHLAVTVGE-TNRR 34
W+KN S K P P GHL + + TNRR
Sbjct: 148 WKKN*NSRWKFPGIAPWGHLMTLLPQITNRR 240
>TC16267 similar to GB|AAP68282.1|31711852|BT008843 At1g73060 {Arabidopsis
thaliana;}, partial (21%)
Length = 418
Score = 25.4 bits (54), Expect = 2.9
Identities = 10/18 (55%), Positives = 15/18 (82%)
Frame = -1
Query: 74 LFEDILLSLGGGTVARRS 91
L+ED+LL GG +++RRS
Sbjct: 343 LYEDMLLGKGGKSISRRS 290
>TC14204 homologue to UP|Q9AVG8 (Q9AVG8) Isopentenyl diphosphate isomerase
1, partial (79%)
Length = 1217
Score = 24.3 bits (51), Expect = 6.6
Identities = 15/37 (40%), Positives = 20/37 (53%), Gaps = 1/37 (2%)
Frame = +1
Query: 41 YLNHPVLQELLDQAYEGYGFNKSGP-LSIPCDEFLFE 76
Y+N L+ELL +A G G K P + D FLF+
Sbjct: 691 YVNREELKELLRKADAGEGGLKLSPWFRLVVDNFLFK 801
>BP085090
Length = 278
Score = 23.9 bits (50), Expect = 8.6
Identities = 9/32 (28%), Positives = 18/32 (56%)
Frame = -2
Query: 48 QELLDQAYEGYGFNKSGPLSIPCDEFLFEDIL 79
Q LL++AY+ +GF + C F++++
Sbjct: 193 QMLLEKAYDEFGFEPRNGRVVLCSVSAFQEVV 98
>TC11248 homologue to UP|P93323 (P93323) Cdc2MsF protein, partial (72%)
Length = 747
Score = 23.9 bits (50), Expect = 8.6
Identities = 10/28 (35%), Positives = 19/28 (67%)
Frame = -1
Query: 47 LQELLDQAYEGYGFNKSGPLSIPCDEFL 74
L +L+ QA++G+G++ L+ +EFL
Sbjct: 402 LAKLVHQAFDGFGWDILSSLTESANEFL 319
>TC18806 similar to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A ligase ,
partial (26%)
Length = 529
Score = 23.9 bits (50), Expect = 8.6
Identities = 16/52 (30%), Positives = 24/52 (45%), Gaps = 1/52 (1%)
Frame = +1
Query: 56 EGYGFNKSGPLSIPCDEFLFEDILLSLGG-GTVARRSSSPVLTKKLDLSFLK 106
+GYG ++GPL+I F E G GTV R + ++ + S K
Sbjct: 265 QGYGMTEAGPLAISL-AFAKEPFKTKSGACGTVVRNAEMKIVDTETGASLPK 417
>TC14695 similar to UP|Q9FQE5 (Q9FQE5) Glutathione S-transferase GST 13 ,
partial (97%)
Length = 1338
Score = 23.9 bits (50), Expect = 8.6
Identities = 12/38 (31%), Positives = 22/38 (57%)
Frame = +3
Query: 26 VTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKS 63
+TV + + F +++NHPV++E L E + F K+
Sbjct: 606 LTVEKFPKLFKWSQEFVNHPVIKEGLPPRDELFAFFKA 719
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,138,724
Number of Sequences: 28460
Number of extensions: 24583
Number of successful extensions: 111
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 110
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 110
length of query: 125
length of database: 4,897,600
effective HSP length: 80
effective length of query: 45
effective length of database: 2,620,800
effective search space: 117936000
effective search space used: 117936000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 50 (23.9 bits)
Medicago: description of AC139525.10


