

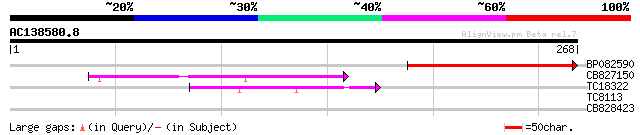
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138580.8 - phase: 0
(268 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP082590 152 6e-38
CB827150 76 7e-15
TC18322 similar to GB|AAP13390.1|30023714|BT006282 At5g44170 {Ar... 47 4e-06
TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter li... 28 1.2
CB828423 28 2.1
>BP082590
Length = 364
Score = 152 bits (384), Expect = 6e-38
Identities = 74/80 (92%), Positives = 78/80 (97%)
Frame = -1
Query: 189 ILGSDVVYSEGAVVDLLETLGQLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTIGRVDQ 248
++GSDVVYSE AVVDL+ETLGQLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTIGRVDQ
Sbjct: 358 VVGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTIGRVDQ 179
Query: 249 TLWHPDYHSNRVVLYVLVKK 268
TLWHP+Y SNRVVLYVLVKK
Sbjct: 178 TLWHPEYRSNRVVLYVLVKK 119
>CB827150
Length = 553
Score = 75.9 bits (185), Expect = 7e-15
Identities = 42/128 (32%), Positives = 72/128 (55%), Gaps = 5/128 (3%)
Frame = +1
Query: 38 LLWG--IQQPTLSKPNAFVAQSSLQLRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIV 95
L WG + L + + ++ ++ L+ GH L Q P+S G+ +WD+ +V
Sbjct: 148 LYWGFFLADCDLMELDRLNTPTTFEMPLEVLGHELQFSQDPNSKH----LGTTVWDASLV 315
Query: 96 LGKFLEHSVDSGMLV---LQGKKIVELGSGCGLVGCIAALLGGEVILTDLPDRMRLLRKN 152
KFLE + G L+GK+++ELG+GCG+ G ALLG +V++TD + + LL++N
Sbjct: 316 FAKFLERNCRKGRFSPAKLKGKRVIELGAGCGVSGFGMALLGCDVVVTDQKEVLPLLQRN 495
Query: 153 IETNMKHI 160
+E N+ +
Sbjct: 496 VERNVSRV 519
>TC18322 similar to GB|AAP13390.1|30023714|BT006282 At5g44170 {Arabidopsis
thaliana;}, partial (41%)
Length = 427
Score = 46.6 bits (109), Expect = 4e-06
Identities = 31/100 (31%), Positives = 51/100 (51%), Gaps = 10/100 (10%)
Frame = +3
Query: 86 GSVMWDSGIVLGKFLEHSVDSG---------MLVLQGKKIVELGSGCGLVGCIAALLG-G 135
G+ +W ++L KF E + +L GK+ VELG GCG+ G LLG
Sbjct: 108 GTSVWPCSLILVKFAERWAQTPPETPNPYTHLLNFAGKRAVELGCGCGVAGMGLYLLGLT 287
Query: 136 EVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTWGD 175
+++LTD+ M L++N++ N LR ++ + L W +
Sbjct: 288 DLVLTDIAPVMPALKRNLKVNKP--LLRKNLKYSILYWNN 401
>TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter like
protein, partial (93%)
Length = 1865
Score = 28.5 bits (62), Expect = 1.2
Identities = 14/42 (33%), Positives = 24/42 (56%)
Frame = +2
Query: 90 WDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVGCIAA 131
+D+G++ G L D G+ Q + + + + C LVGC+AA
Sbjct: 164 YDTGVMSGALLFIKEDIGISDTQQEVLAGILNICALVGCLAA 289
>CB828423
Length = 512
Score = 27.7 bits (60), Expect = 2.1
Identities = 14/41 (34%), Positives = 23/41 (55%)
Frame = +1
Query: 91 DSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVGCIAA 131
D+G++ G L D G+ Q + + + + C LVGC+AA
Sbjct: 370 DTGVMSGALLFIKEDIGISDTQQEVLAGILNICALVGCLAA 492
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.136 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,211,568
Number of Sequences: 28460
Number of extensions: 52426
Number of successful extensions: 243
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 241
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 242
length of query: 268
length of database: 4,897,600
effective HSP length: 89
effective length of query: 179
effective length of database: 2,364,660
effective search space: 423274140
effective search space used: 423274140
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 54 (25.4 bits)
Medicago: description of AC138580.8


