

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138451.12 - phase: 0
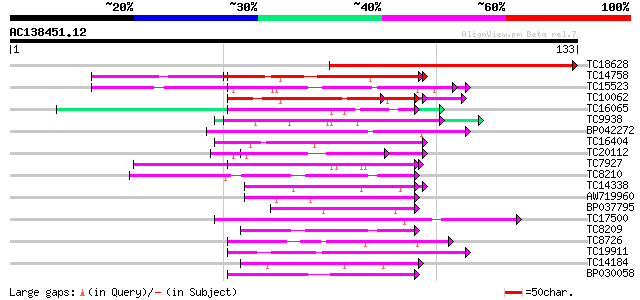
(133 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC18628 weakly similar to PIR|T09527|T09527 glycine-rich protein... 77 8e-16
TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding p... 58 6e-10
TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-bindi... 51 8e-08
TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein... 48 7e-07
TC16065 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich... 44 1e-05
TC9938 weakly similar to GB|AAM97131.1|22531255|AY136466 ribonuc... 42 3e-05
BP042272 42 4e-05
TC16404 similar to UP|P93320 (P93320) Cdc2MsC protein, partial (... 42 5e-05
TC20112 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich... 41 6e-05
TC7927 similar to UP|Q93YR3 (Q93YR3) HSP associated protein like... 41 8e-05
TC8210 homologue to PIR|T10586|T10586 small nuclear ribonucleopr... 40 1e-04
TC14338 similar to UP|Q9MAH5 (Q9MAH5) F12M16.16, partial (33%) 40 1e-04
AW719960 40 2e-04
BP037795 39 2e-04
TC17500 UP|Q9ZPL9 (Q9ZPL9) Nodule-enhanced protein phosphatase t... 39 2e-04
TC8209 homologue to PIR|T10586|T10586 small nuclear ribonucleopr... 39 3e-04
TC8726 similar to GB|AAO24533.1|27808506|BT003101 At2g41420 {Ara... 39 3e-04
TC19911 weakly similar to PIR|A44805|A44805 eggshell protein pre... 38 5e-04
TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-l... 38 7e-04
BP030058 38 7e-04
>TC18628 weakly similar to PIR|T09527|T09527 glycine-rich protein 1 -
chickpea {Cicer arietinum;} , partial (32%)
Length = 424
Score = 77.4 bits (189), Expect = 8e-16
Identities = 34/58 (58%), Positives = 38/58 (64%)
Frame = +3
Query: 76 GGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAGEAVAMQTENNTRN 133
GG GG GGG G GG YCR+GCC Y GSCR+CC YAGEAVA QT + T+N
Sbjct: 6 GGGGRGHGGRGGGGGRGGGGGGYCRHGCCDHGYRGSCRRCCHYAGEAVAAQTADKTQN 179
>TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding protein 2,
partial (93%)
Length = 725
Score = 57.8 bits (138), Expect = 6e-10
Identities = 36/83 (43%), Positives = 43/83 (51%), Gaps = 4/83 (4%)
Frame = +1
Query: 20 SSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYP 79
+SE + R E + E D + +N+A+ GGG GRGGGG YGGG R GGY
Sbjct: 229 TSEEAMRSAIEGMNGN--ELDGRNITVNEAQARGGGGGGGGRGGGG-YGGGGGRREGGYG 399
Query: 80 GNRG----GNYGGGYPGNRGGNY 98
G G G GGGY G GG Y
Sbjct: 400 GGGGSRGYGGGGGGYGGGGGGGY 468
Score = 49.3 bits (116), Expect = 2e-07
Identities = 26/48 (54%), Positives = 29/48 (60%)
Frame = +1
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
YG GGGY G GGGG YGG R GGY G+ GG+ G G GGN+
Sbjct: 421 YGGGGGGYGG-GGGGGYGG---RREGGYGGDGGGSRYGSGGGGGGGNW 552
Score = 47.4 bits (111), Expect = 9e-07
Identities = 26/49 (53%), Positives = 28/49 (57%), Gaps = 3/49 (6%)
Frame = +1
Query: 52 GSYGGGYPGRG---GGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGN 97
G YGGG RG GGG YGGG GGY G R G YGG G+R G+
Sbjct: 388 GGYGGGGGSRGYGGGGGGYGGG---GGGGYGGRREGGYGGDGGGSRYGS 525
>TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-binding,
abscisic acid-inducible protein, complete
Length = 972
Score = 50.8 bits (120), Expect = 8e-08
Identities = 40/100 (40%), Positives = 50/100 (50%), Gaps = 11/100 (11%)
Frame = +2
Query: 20 SSEVSARDLTEASSNTQKEADDQTNELNDAKY-----GSYGGGYPG-RGGGGNYGGGYPR 73
+SE S +D E + ++ D + +N+A+ G GGGY G R GG GGG R
Sbjct: 503 ASEQSMKDAIEGMNG--QDIDGRNVTVNEAQARGRGGGGGGGGYGGGRREGGYGGGGGSR 676
Query: 74 NRGGYPGNRGGNYGGGYPGNR---GGNY--CRYGCCGDRY 108
GG G GG+ GGGY G R GG Y R G G RY
Sbjct: 677 YSGGGGGGYGGSGGGGYGGRREGGGGGYGGSREGGGGSRY 796
Score = 46.6 bits (109), Expect = 1e-06
Identities = 30/58 (51%), Positives = 31/58 (52%), Gaps = 4/58 (6%)
Frame = +2
Query: 52 GSYGGGYPGR---GGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNR-GGNYCRYGCCG 105
G YGGG R GGGG YGG GGY G R G GGGY G+R GG RY G
Sbjct: 647 GGYGGGGGSRYSGGGGGGYGGS---GGGGYGGRREGG-GGGYGGSREGGGGSRYSSGG 808
Score = 37.0 bits (84), Expect = 0.001
Identities = 22/46 (47%), Positives = 26/46 (55%), Gaps = 2/46 (4%)
Frame = +2
Query: 55 GGGYPGRG--GGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
GGG G G GGG YGG GGY G+R G GGG + GG++
Sbjct: 683 GGGGGGYGGSGGGGYGGRREGGGGGYGGSREG--GGGSRYSSGGSW 814
>TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein 2, partial
(66%)
Length = 481
Score = 47.8 bits (112), Expect = 7e-07
Identities = 27/45 (60%), Positives = 27/45 (60%)
Frame = +3
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G YGGG GRGGGG GGG GGY G GG GGG G RGG
Sbjct: 342 GGYGGG--GRGGGGYGGGGGYGGSGGY-GGGGGYGGGGRGGGRGG 467
Score = 46.2 bits (108), Expect = 2e-06
Identities = 33/59 (55%), Positives = 33/59 (55%), Gaps = 3/59 (5%)
Frame = +3
Query: 52 GSYGGGYPGRG-GGGNYGGGYPRNRGGYPGNRGGNYG--GGYPGNRGGNYCRYGCCGDR 107
G GGGY G G GGG YGGG R GGY G GG YG GGY G GG Y G G R
Sbjct: 300 GRGGGGYGGGGYGGGGYGGG-GRGGGGYGG--GGGYGGSGGYGG--GGGYGGGGRGGGR 461
Score = 44.3 bits (103), Expect = 7e-06
Identities = 28/47 (59%), Positives = 28/47 (59%)
Frame = +3
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
G GGG GRGGGG GGGY GGY G GG GGGY G GG Y
Sbjct: 282 GGAGGG--GRGGGGYGGGGY--GGGGYGG--GGRGGGGYGG--GGGY 398
Score = 41.2 bits (95), Expect = 6e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Frame = +3
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGG 88
G YGGG G GG G YGGG GG G RGG GG
Sbjct: 372 GGYGGG-GGYGGSGGYGGGGGYGGGGRGGGRGGGGGG 479
Score = 26.2 bits (56), Expect = 2.0
Identities = 17/39 (43%), Positives = 17/39 (43%)
Frame = +3
Query: 82 RGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAG 120
RGG GGG G GG Y G G Y G R Y G
Sbjct: 276 RGGGAGGG--GRGGGGYGGGGYGGGGYGGGGRGGGGYGG 386
Score = 25.8 bits (55), Expect = 2.7
Identities = 14/21 (66%), Positives = 14/21 (66%), Gaps = 1/21 (4%)
Frame = +3
Query: 51 YGSYGGGYPGRG-GGGNYGGG 70
YG GGGY G G GGG GGG
Sbjct: 414 YGG-GGGYGGGGRGGGRGGGG 473
>TC16065 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich
glycoprotein DZ-HRGP precursor, partial (20%)
Length = 608
Score = 43.5 bits (101), Expect = 1e-05
Identities = 29/92 (31%), Positives = 37/92 (39%), Gaps = 1/92 (1%)
Frame = -1
Query: 12 LLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGGGNYGGGY 71
LL I++ E S + +++ + GGG PG GG YGG
Sbjct: 500 LLGFIMVRKRRTDGPSGGEGSEGQHSHSTERSAPPPQQAAAAPGGGGPGPQGGRGYGGPP 321
Query: 72 PRNRGG-YPGNRGGNYGGGYPGNRGGNYCRYG 102
RGG Y G G YGGG G G +YG
Sbjct: 320 QGGRGGGYGGGGGPGYGGGGRGGHGVPQQQYG 225
Score = 42.0 bits (97), Expect = 4e-05
Identities = 28/66 (42%), Positives = 29/66 (43%), Gaps = 21/66 (31%)
Frame = -1
Query: 52 GSYGGGYPGRGGGGNYGGGYPRN---------------------RGGYPGNRGGNYGGGY 90
G GGG PG GGGG G G P+ RGGY G GG YGGG
Sbjct: 302 GYGGGGGPGYGGGGRGGHGVPQQQYGGPPEYQGRGRGGPSQQGGRGGYSGG-GGGYGGG- 129
Query: 91 PGNRGG 96
G RGG
Sbjct: 128 -GGRGG 114
Score = 35.0 bits (79), Expect = 0.004
Identities = 19/39 (48%), Positives = 21/39 (53%)
Frame = -1
Query: 56 GGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNR 94
GGY G GGG YGGG R GG RG + G P +R
Sbjct: 167 GGYSG--GGGGYGGGGGRGGGGMGSGRGVSPSHGGPPSR 57
Score = 31.2 bits (69), Expect = 0.063
Identities = 15/33 (45%), Positives = 17/33 (51%)
Frame = -1
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGG 84
G Y GG G GGGG GGG + G + GG
Sbjct: 167 GGYSGGGGGYGGGGGRGGGGMGSGRGVSPSHGG 69
Score = 29.6 bits (65), Expect = 0.18
Identities = 20/50 (40%), Positives = 21/50 (42%), Gaps = 4/50 (8%)
Frame = -1
Query: 50 KYGSYGGGYP----GRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRG 95
+Y G G P GRGG GGGY G GG GGG RG
Sbjct: 215 EYQGRGRGGPSQQGGRGGYSGGGGGY--------GGGGGRGGGGMGSGRG 90
Score = 26.9 bits (58), Expect = 1.2
Identities = 13/28 (46%), Positives = 14/28 (49%)
Frame = -1
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYP 79
G YGGG GGG G G + GG P
Sbjct: 146 GGYGGGGGRGGGGMGSGRGVSPSHGGPP 63
>TC9938 weakly similar to GB|AAM97131.1|22531255|AY136466
ribonucleoprotein-like {Arabidopsis thaliana;}, partial
(9%)
Length = 633
Score = 42.4 bits (98), Expect = 3e-05
Identities = 24/56 (42%), Positives = 28/56 (49%), Gaps = 4/56 (7%)
Frame = +1
Query: 51 YGSYGGGYPGRGGG---GNYGGGYPRN-RGGYPGNRGGNYGGGYPGNRGGNYCRYG 102
YG+YG G GG G Y GGY + GGY G GG++ G G G Y YG
Sbjct: 190 YGAYGAGAAAGGGSSAAGAYQGGYDASLAGGYGGASGGSFYGSRGGYGAGRYHPYG 357
Score = 37.4 bits (85), Expect = 9e-04
Identities = 28/92 (30%), Positives = 36/92 (38%), Gaps = 29/92 (31%)
Frame = +1
Query: 49 AKYGSYGG---------GYPGRGGGGNYGGGYPR--NRGGYPG----------------- 80
+++G YGG G P G G YGG + R + GGY G
Sbjct: 49 SEFGGYGGYAGAMGPYRGDPSLGFAGRYGGSFSRGYDLGGYGGASESYGAYGAGAAAGGG 228
Query: 81 -NRGGNYGGGYPGNRGGNYCRYGCCGDRYYGS 111
+ G Y GGY + G Y G G +YGS
Sbjct: 229 SSAAGAYQGGYDASLAGGY--GGASGGSFYGS 318
Score = 31.6 bits (70), Expect = 0.048
Identities = 22/69 (31%), Positives = 29/69 (41%), Gaps = 4/69 (5%)
Frame = +1
Query: 47 NDAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPG--NRGGNYCRYGCC 104
+ A Y G Y G G GGY G Y G+ + G Y G +RG + YG
Sbjct: 1 SSAAYAGRSGAYGGYGSEFGGYGGYAGAMGPYRGDPSLGFAGRYGGSFSRGYDLGGYGGA 180
Query: 105 GDRY--YGS 111
+ Y YG+
Sbjct: 181 SESYGAYGA 207
>BP042272
Length = 471
Score = 42.0 bits (97), Expect = 4e-05
Identities = 24/63 (38%), Positives = 33/63 (52%), Gaps = 1/63 (1%)
Frame = +1
Query: 47 NDAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRG-GNYCRYGCCG 105
+D + G + R GG GGG RGG+ +RGG +GGG + G G+ R G G
Sbjct: 196 DDGPASDFYGKFSDRDGGSRRGGGSRDRRGGFKSSRGG-WGGGRDSDDGFGDQYRRGGGG 372
Query: 106 DRY 108
D+Y
Sbjct: 373 DQY 381
Score = 28.5 bits (62), Expect = 0.41
Identities = 16/45 (35%), Positives = 17/45 (37%)
Frame = +1
Query: 53 SYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGN 97
S GG GR +G Y R GG RGG GGN
Sbjct: 298 SRGGWGGGRDSDDGFGDQYRRGGGGDQYRRGGGSSSSRNSRTGGN 432
>TC16404 similar to UP|P93320 (P93320) Cdc2MsC protein, partial (15%)
Length = 584
Score = 41.6 bits (96), Expect = 5e-05
Identities = 26/55 (47%), Positives = 28/55 (50%), Gaps = 5/55 (9%)
Frame = +2
Query: 49 AKYGSYG--GGYPGRGGGGNYGGG---YPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
A YGS G GG P RGGGG YG G YP+ G Y G+ G GNR Y
Sbjct: 71 APYGSSGMPGGGP-RGGGGGYGAGAPNYPQQGGPYGGSAAGRGSNMMGGNRNQQY 232
Score = 35.0 bits (79), Expect = 0.004
Identities = 24/56 (42%), Positives = 26/56 (45%), Gaps = 9/56 (16%)
Frame = +2
Query: 52 GSYGGG------YPGRGGGGNYG-GGYPRNRGGYPGNRGGNYGGGYPG--NRGGNY 98
G GGG YP +G G YG G P GG P GG YG G P +GG Y
Sbjct: 14 GXQGGGGYGSGPYPPQGRGAPYGSSGMP---GGGPRGGGGGYGAGAPNYPQQGGPY 172
Score = 31.6 bits (70), Expect = 0.048
Identities = 19/48 (39%), Positives = 20/48 (41%)
Frame = +2
Query: 61 RGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRY 108
+GGGG G YP G P G GGG G GG YG Y
Sbjct: 20 QGGGGYGSGPYPPQGRGAPYGSSGMPGGGPRGGGGG----YGAGAPNY 151
>TC20112 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich
glycoprotein DZ-HRGP precursor, partial (11%)
Length = 523
Score = 41.2 bits (95), Expect = 6e-05
Identities = 20/44 (45%), Positives = 23/44 (51%)
Frame = +2
Query: 55 GGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
GGG+ GRGG Y G GG G GG+ GG G RGG +
Sbjct: 365 GGGFGGRGGDRKYDTGRYSGGGGGRGRGGGSIRGGGRGGRGGGF 496
Score = 39.7 bits (91), Expect = 2e-04
Identities = 25/46 (54%), Positives = 26/46 (56%), Gaps = 4/46 (8%)
Frame = +2
Query: 48 DAKY--GSY--GGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGG 89
D KY G Y GGG GRGGG GGG RGG G GG +GGG
Sbjct: 392 DRKYDTGRYSGGGGGRGRGGGSIRGGG----RGGRGGGFGGRHGGG 517
Score = 24.3 bits (51), Expect = 7.7
Identities = 9/15 (60%), Positives = 11/15 (73%)
Frame = +2
Query: 52 GSYGGGYPGRGGGGN 66
G GGG+ GR GGG+
Sbjct: 476 GGRGGGFGGRHGGGS 520
>TC7927 similar to UP|Q93YR3 (Q93YR3) HSP associated protein like, partial
(56%)
Length = 1116
Score = 40.8 bits (94), Expect = 8e-05
Identities = 24/50 (48%), Positives = 26/50 (52%), Gaps = 4/50 (8%)
Frame = +1
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNR--GGYPGNR--GGNYGGGYPGNRGGN 97
G GGG PG GG GG+P GG+PG GG GGG PG GN
Sbjct: 511 GFPGGGMPGGFPGGGMPGGFPGGGMPGGFPGGGMPGGFPGGGMPGGVPGN 660
Score = 39.3 bits (90), Expect = 2e-04
Identities = 25/71 (35%), Positives = 31/71 (43%), Gaps = 4/71 (5%)
Frame = +1
Query: 30 EASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGGGNYGGGYPRN--RGGYPGN--RGGN 85
+ S++ + E G GGG PG GG GG+P GG+PG GG
Sbjct: 391 QEQSSSSRNPGGMPGEFPGMPGGFPGGGMPGGFPGGGMPGGFPGGGMPGGFPGGGMPGGF 570
Query: 86 YGGGYPGNRGG 96
GGG PG G
Sbjct: 571 PGGGMPGGFPG 603
>TC8210 homologue to PIR|T10586|T10586 small nuclear
ribonucleoprotein-associated protein homolog
F9F13.90 - Arabidopsis thaliana {Arabidopsis
thaliana;}, partial (16%)
Length = 698
Score = 40.4 bits (93), Expect = 1e-04
Identities = 28/74 (37%), Positives = 35/74 (46%), Gaps = 6/74 (8%)
Frame = -3
Query: 29 TEASSNTQKEADDQTNELNDA------KYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNR 82
T S + + ++TN +D ++G GGG PGRGG G P GG PG
Sbjct: 339 TSPMSISSEHQQEETNPFHDHCCC*F*EFG--GGGMPGRGGA*T---GAPPGGGGIPGRG 175
Query: 83 GGNYGGGYPGNRGG 96
GG G G PG GG
Sbjct: 174 GG--GPGIPGRGGG 139
Score = 31.6 bits (70), Expect = 0.048
Identities = 24/54 (44%), Positives = 24/54 (44%), Gaps = 6/54 (11%)
Frame = -3
Query: 55 GGGYPGRGGGGNYGGGYPRNRGG------YPGNRGGNYGGGYPGNRGGNYCRYG 102
GGG PGRGGG G G P GG PG GG G P G YC G
Sbjct: 198 GGGIPGRGGG---GPGIPGRGGGGGPLIICPG--GGGPIGLCPNCGGTPYCGGG 52
Score = 31.2 bits (69), Expect = 0.063
Identities = 24/55 (43%), Positives = 24/55 (43%), Gaps = 11/55 (20%)
Frame = -3
Query: 52 GSYGGGYPGRGGGGN-----YGGGYP----RNRGGYPGNRGG--NYGGGYPGNRG 95
G G G PGRGGGG GGG P N GG P GG G PG G
Sbjct: 177 GGGGPGIPGRGGGGGPLIICPGGGGPIGLCPNCGGTPYCGGGPPGLGAKLPGGIG 13
>TC14338 similar to UP|Q9MAH5 (Q9MAH5) F12M16.16, partial (33%)
Length = 1751
Score = 40.0 bits (92), Expect = 1e-04
Identities = 22/44 (50%), Positives = 23/44 (52%), Gaps = 1/44 (2%)
Frame = +2
Query: 56 GGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGY-PGNRGGNY 98
GGYP GN G P N GGYP N N GGY P N+ G Y
Sbjct: 869 GGYPASPRPGNMPGPPPSNPGGYPPN---NQAGGYHPQNQAGGY 991
Score = 40.0 bits (92), Expect = 1e-04
Identities = 22/45 (48%), Positives = 24/45 (52%), Gaps = 4/45 (8%)
Frame = +2
Query: 56 GGYPGRGGGG---NYGGGYPRNRGGYPGN-RGGNYGGGYPGNRGG 96
GG+P GG N GG P N GGYP + R GN G P N GG
Sbjct: 800 GGFPPNNAGGPPSNQGGFPPNNAGGYPASPRPGNMPGPPPSNPGG 934
Score = 37.0 bits (84), Expect = 0.001
Identities = 23/70 (32%), Positives = 30/70 (42%), Gaps = 3/70 (4%)
Frame = +2
Query: 30 EASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGG--GNYGGGYPRNRGGYPGNRGGNY- 86
+ S N + ++ N + GG P GG N GG P N+GG+P N G Y
Sbjct: 701 DRSRNFDRRRENVVNRDYQNRPPPNAGGPPSNQGGFPPNNAGGPPSNQGGFPPNNAGGYP 880
Query: 87 GGGYPGNRGG 96
PGN G
Sbjct: 881 ASPRPGNMPG 910
Score = 31.6 bits (70), Expect = 0.048
Identities = 20/52 (38%), Positives = 21/52 (39%), Gaps = 6/52 (11%)
Frame = +2
Query: 53 SYGGGYPGRGGG------GNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
S G YP G NY G P N GG P N GYP N+ NY
Sbjct: 1055 SQAGVYPPNNQGVYPPNQNNYAGRTPPNVGGAPPNSAYGPNAGYPSNQ-NNY 1207
Score = 25.0 bits (53), Expect = 4.5
Identities = 20/46 (43%), Positives = 20/46 (43%), Gaps = 4/46 (8%)
Frame = -3
Query: 56 GGYPGRGGGGNYGGGYPRNRGGY-PGNRGGN---YGGGYPGNRGGN 97
GGYP GG G R GY P GGN GG P GGN
Sbjct: 942 GGYPPGLLGGGPGMLPGRGEAGYPPALLGGNPP*LLGGPPALLGGN 805
>AW719960
Length = 449
Score = 39.7 bits (91), Expect = 2e-04
Identities = 23/49 (46%), Positives = 24/49 (48%), Gaps = 8/49 (16%)
Frame = +3
Query: 56 GGYPGR----GGGGNYGG----GYPRNRGGYPGNRGGNYGGGYPGNRGG 96
GGYPG+ G G Y G GYP GYPG G GYPG GG
Sbjct: 162 GGYPGQYGAPGAPGAYPGAPPAGYPGAPAGYPGAYPGAPAAGYPGAPGG 308
Score = 37.0 bits (84), Expect = 0.001
Identities = 23/51 (45%), Positives = 26/51 (50%), Gaps = 5/51 (9%)
Frame = +3
Query: 52 GSYGG----GYPGRGGGGNYGGGYP-RNRGGYPGNRGGNYGGGYPGNRGGN 97
G+Y G GYPG G Y G YP GYPG GG G G PG+ G+
Sbjct: 201 GAYPGAPPAGYPGAPAG--YPGAYPGAPAAGYPGAPGGAPGYGTPGHSAGS 347
>BP037795
Length = 548
Score = 39.3 bits (90), Expect = 2e-04
Identities = 22/42 (52%), Positives = 23/42 (54%), Gaps = 7/42 (16%)
Frame = -3
Query: 62 GGGGNYGGGYP--RNRGGYPGNRGGNYGGG-----YPGNRGG 96
GG GN GGGY N GYP GG+ GG Y GNRGG
Sbjct: 435 GGPGNNGGGYRPNNNASGYPPRDGGHGRGGRPQNNYAGNRGG 310
Score = 27.7 bits (60), Expect = 0.70
Identities = 13/30 (43%), Positives = 15/30 (49%), Gaps = 5/30 (16%)
Frame = -3
Query: 73 RNRGGYPGNRGGNY-----GGGYPGNRGGN 97
R+ G PGN GG Y GYP GG+
Sbjct: 447 RDNAGGPGNNGGGYRPNNNASGYPPRDGGH 358
>TC17500 UP|Q9ZPL9 (Q9ZPL9) Nodule-enhanced protein phosphatase type 2C,
complete
Length = 1325
Score = 39.3 bits (90), Expect = 2e-04
Identities = 26/74 (35%), Positives = 33/74 (44%), Gaps = 2/74 (2%)
Frame = +2
Query: 49 AKYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYP--GNRGGNYCRYGCCGD 106
++ S GGG PG GGG G G GG+ G + G GG C G
Sbjct: 500 SRRSSGGGGVPGAAVSACGGGG*EVREWGGGGGLGGSDGRVFS*YGR*GGG*C--GVEDG 673
Query: 107 RYYGSCRKCCSYAG 120
++G CR+CCS G
Sbjct: 674 GFHGGCRRCCSGGG 715
>TC8209 homologue to PIR|T10586|T10586 small nuclear
ribonucleoprotein-associated protein homolog
F9F13.90 - Arabidopsis thaliana {Arabidopsis
thaliana;}, partial (19%)
Length = 635
Score = 38.9 bits (89), Expect = 3e-04
Identities = 22/42 (52%), Positives = 22/42 (52%)
Frame = -2
Query: 55 GGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
GGG PGRGG G P GG PG GG G G PG GG
Sbjct: 211 GGGMPGRGGA*T---GAPPGGGGIPGRGGG--GPGIPGRGGG 101
Score = 31.6 bits (70), Expect = 0.048
Identities = 24/54 (44%), Positives = 24/54 (44%), Gaps = 6/54 (11%)
Frame = -2
Query: 55 GGGYPGRGGGGNYGGGYPRNRGG------YPGNRGGNYGGGYPGNRGGNYCRYG 102
GGG PGRGGG G G P GG PG GG G P G YC G
Sbjct: 160 GGGIPGRGGG---GPGIPGRGGGGGPLIICPG--GGGPIGLCPNCGGTPYCGGG 14
Score = 30.8 bits (68), Expect = 0.082
Identities = 23/50 (46%), Positives = 23/50 (46%), Gaps = 9/50 (18%)
Frame = -2
Query: 52 GSYGGGYPGRGGGGN-----YGGGYP----RNRGGYPGNRGGNYGGGYPG 92
G G G PGRGGGG GGG P N GG P GGG PG
Sbjct: 139 GGGGPGIPGRGGGGGPLIICPGGGGPIGLCPNCGGTP-----YCGGGPPG 5
>TC8726 similar to GB|AAO24533.1|27808506|BT003101 At2g41420 {Arabidopsis
thaliana;}, partial (95%)
Length = 820
Score = 38.9 bits (89), Expect = 3e-04
Identities = 29/64 (45%), Positives = 30/64 (46%), Gaps = 11/64 (17%)
Frame = -2
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNR---GGNYGGGYPGNR--------GGNYCR 100
G+YGGG P G GG GGYP GGYPG GG GGYP GG
Sbjct: 303 GAYGGG*PCAGYGG---GGYPC-CGGYPGGGYP*GGYPCGGYPSFPYPSGG*P*GGGTPT 136
Query: 101 YGCC 104
GCC
Sbjct: 135 GGCC 124
>TC19911 weakly similar to PIR|A44805|A44805 eggshell protein precursor -
fluke (Schistosoma haematobium) {Schistosoma
haematobium;} , partial (24%)
Length = 350
Score = 38.1 bits (87), Expect = 5e-04
Identities = 24/57 (42%), Positives = 27/57 (47%)
Frame = +1
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRY 108
G GGGY G GG+ GGY + GG G GG YG G + G Y G G Y
Sbjct: 163 GRTGGGY---GDGGHSAGGYG-DHGGRSGGGGGGYGDGGLSSGSGGYGDGGRSGVGY 321
Score = 34.7 bits (78), Expect = 0.006
Identities = 24/54 (44%), Positives = 28/54 (51%), Gaps = 6/54 (11%)
Frame = +1
Query: 51 YGSYGGGYP------GRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
+G GGGY GR GGG GG+ + GGY G+ GG GGG GG Y
Sbjct: 118 HGLSGGGYDDDNRSGGRTGGGYGDGGH--SAGGY-GDHGGRSGGG-----GGGY 255
Score = 34.3 bits (77), Expect = 0.007
Identities = 22/62 (35%), Positives = 28/62 (44%)
Frame = +1
Query: 41 DQTNELNDAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCR 100
D N G YG G GG G++GG GGY G+ G + G G G+ G +
Sbjct: 142 DDDNRSGGRTGGGYGDGGHSAGGYGDHGGRSGGGGGGY-GDGGLSSGSGGYGDGGRSGVG 318
Query: 101 YG 102
YG
Sbjct: 319 YG 324
Score = 33.5 bits (75), Expect = 0.013
Identities = 20/42 (47%), Positives = 20/42 (47%), Gaps = 4/42 (9%)
Frame = +1
Query: 52 GSYG--GGYPGRGGGGNYGGGYPRNRGGY--PGNRGGNYGGG 89
G YG GG G GGGG GG GGY G G YG G
Sbjct: 205 GGYGDHGGRSGGGGGGYGDGGLSSGSGGYGDGGRSGVGYGDG 330
Score = 30.8 bits (68), Expect = 0.082
Identities = 23/59 (38%), Positives = 28/59 (46%), Gaps = 3/59 (5%)
Frame = +1
Query: 47 NDAKYGS--YGGGYPGRGGGGNYGGGY-PRNRGGYPGNRGGNYGGGYPGNRGGNYCRYG 102
ND ++ +GG G G GGGY NR G G GG YG G G+ G Y +G
Sbjct: 61 NDHRHSDNEHGGRRSETTGHGLSGGGYDDDNRSG--GRTGGGYGDG--GHSAGGYGDHG 225
>TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-like
protein, partial (57%)
Length = 1141
Score = 37.7 bits (86), Expect = 7e-04
Identities = 24/52 (46%), Positives = 26/52 (49%), Gaps = 9/52 (17%)
Frame = +3
Query: 55 GGGYP--GRGGGGNYGGGYPRNRGG----YPGNRGGNY---GGGYPGNRGGN 97
GGGY G GGG + GGGY GG G GG Y GGGY + GN
Sbjct: 366 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGN 521
Score = 35.8 bits (81), Expect = 0.003
Identities = 32/96 (33%), Positives = 36/96 (37%), Gaps = 13/96 (13%)
Frame = +3
Query: 20 SSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGGGNYGGGYPR------ 73
+ E S D + E E K SY P GGGG Y GY +
Sbjct: 180 TDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGG-YDSGYNKTAYSTD 356
Query: 74 --NRGGY-PGNRGGNY--GGGY--PGNRGGNYCRYG 102
+ GGY G GG Y GGGY G GG Y G
Sbjct: 357 EPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGG 464
Score = 35.0 bits (79), Expect = 0.004
Identities = 24/46 (52%), Positives = 24/46 (52%), Gaps = 4/46 (8%)
Frame = +3
Query: 53 SYGGGY-PGRGGGGNY---GGGYPRNRGGYPGNRGGNYGGGYPGNR 94
S GGGY G GGGG Y GGG GGY GG YG GNR
Sbjct: 405 STGGGYGTGGGGGGEYSTGGGG-----GGY-STGGGGYGNDDSGNR 524
>BP030058
Length = 455
Score = 37.7 bits (86), Expect = 7e-04
Identities = 21/45 (46%), Positives = 23/45 (50%)
Frame = +1
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G +GGG G GGGG GGG GG G+ G GG G GG
Sbjct: 289 GYFGGGGEGGGGGGGDGGG-----GGGDGHGSGGGGGEGDGGGGG 408
Score = 28.9 bits (63), Expect = 0.31
Identities = 16/41 (39%), Positives = 19/41 (46%)
Frame = +1
Query: 56 GGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G Y GGGG+ G + G + G G GGG G GG
Sbjct: 229 GTYMILGGGGDKGLQFCGQEGYFGGGGEGGGGGGGDGGGGG 351
Score = 25.8 bits (55), Expect = 2.7
Identities = 13/23 (56%), Positives = 15/23 (64%)
Frame = +1
Query: 51 YGSYGGGYPGRGGGGNYGGGYPR 73
+GS GGG G GGG GGG P+
Sbjct: 361 HGSGGGGGEGDGGG---GGGSPQ 420
Score = 24.6 bits (52), Expect = 5.9
Identities = 18/45 (40%), Positives = 19/45 (42%), Gaps = 3/45 (6%)
Frame = +1
Query: 55 GGGYPGR---GGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
GGG G G G +GGG GG G G GGG GG
Sbjct: 250 GGGDKGLQFCGQEGYFGGG---GEGGGGGGGDGGGGGGDGHGSGG 375
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.139 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,402,330
Number of Sequences: 28460
Number of extensions: 43475
Number of successful extensions: 1411
Number of sequences better than 10.0: 274
Number of HSP's better than 10.0 without gapping: 673
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1018
length of query: 133
length of database: 4,897,600
effective HSP length: 80
effective length of query: 53
effective length of database: 2,620,800
effective search space: 138902400
effective search space used: 138902400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 50 (23.9 bits)
Medicago: description of AC138451.12


