

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138199.5 + phase: 0
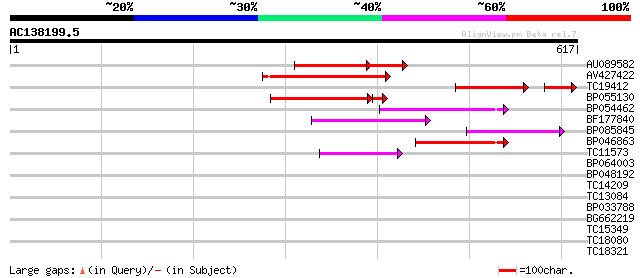
(617 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AU089582 110 2e-41
AV427422 139 1e-33
TC19412 similar to UP|Q84KB0 (Q84KB0) Pol protein, partial (7%) 99 6e-27
BP055130 106 1e-24
BP054462 95 3e-20
BF177840 91 4e-19
BP085845 61 5e-10
BP046863 59 2e-09
TC11573 similar to UP|Q9M6N4 (Q9M6N4) Pol protein integrase regi... 57 1e-08
BP064003 33 0.13
BP048192 32 0.39
TC14209 similar to UP|AAQ72788 (AAQ72788) Hypersensitive-induced... 31 0.67
TC13084 30 1.1
BP033788 30 1.5
BG662219 28 5.7
TC15349 homologue to UP|143C_SOYBN (Q96452) 14-3-3-like protein ... 27 7.4
TC18080 similar to GB|AAL90935.1|19699138|AY090274 AT3g18790/MVE... 27 9.7
TC18321 similar to UP|ORN_ARATH (Q9ZVE0) Probable oligoribonucle... 27 9.7
>AU089582
Length = 383
Score = 110 bits (276), Expect(2) = 2e-41
Identities = 52/83 (62%), Positives = 66/83 (78%)
Frame = +2
Query: 311 AQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSE 370
+Q A+IY+ EIV LHGVP SI+SDR +FTS FW+S Q ALG++L++S+A+HPQTDGQSE
Sbjct: 14 SQYAKIYLDEIVSLHGVPVSIISDRGAQFTSHFWRSFQTALGTRLKMSTAFHPQTDGQSE 193
Query: 371 RTIQSLEDLLRICVLEQGGTWDS 393
RTIQ LED+LR CV + G S
Sbjct: 194 RTIQILEDMLRACVXDLRGXLGS 262
Score = 75.9 bits (185), Expect(2) = 2e-41
Identities = 33/45 (73%), Positives = 37/45 (81%)
Frame = +3
Query: 389 GTWDSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRIPLCWFESG 433
G+WD +L L+EF YNNSY SSI MAPFEALYGRRCR P+ WFE G
Sbjct: 249 GSWDQYLSLMEFAYNNSYRSSI*MAPFEALYGRRCRSPIGWFEVG 383
>AV427422
Length = 417
Score = 139 bits (350), Expect = 1e-33
Identities = 65/139 (46%), Positives = 96/139 (68%)
Frame = +1
Query: 276 TSLPNTPRGNDAIWVIVDRLTKSAHFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDR 335
T LP + +G +A+ V+VDRL+K +HF+P+ + +A+I+++E+V+LHGVP SIVSDR
Sbjct: 4 TGLPKS-KGYEAVLVVVDRLSKFSHFVPLKHPYTAKVIADIFVREVVRLHGVPLSIVSDR 180
Query: 336 DPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHL 395
DP F S FWK L + G+KL++S+AYHP++DGQ+E + LE LR + +Q +W +
Sbjct: 181 DPLFMSNFWKELFKMQGTKLKMSTAYHPESDGQTEVVNRCLETYLRCFIADQPKSWAHWV 360
Query: 396 PLIEFTYNNSYHSSIGMAP 414
P E+ YN SYH S G P
Sbjct: 361 PWAEYWYNTSYHVSTGQTP 417
>TC19412 similar to UP|Q84KB0 (Q84KB0) Pol protein, partial (7%)
Length = 519
Score = 99.0 bits (245), Expect(2) = 6e-27
Identities = 47/79 (59%), Positives = 64/79 (80%)
Frame = +3
Query: 486 RVTPMTGVGRALKSKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHVSQLQKYV 545
RV+PM GV R K KL+P+FIGP+++LERVG+V+YR+ LPP LS +H VFHVS L+KY+
Sbjct: 3 RVSPMKGVLRFGKKGKLSPRFIGPFEVLERVGSVSYRLALPPDLSAVHPVFHVSMLRKYL 182
Query: 546 PDPSHVIQSDDVQVRDNLT 564
DPSHVI+ +DVQ+ +L+
Sbjct: 183 YDPSHVIRHEDVQLDVHLS 239
Score = 39.3 bits (90), Expect(2) = 6e-27
Identities = 16/34 (47%), Positives = 21/34 (61%)
Frame = +2
Query: 583 KEIPLVRVVWTGATGESLTWELESKMLVSYPELF 616
K++ V+V+W G +GE TWE E M YP LF
Sbjct: 296 KDVGSVKVLWRGPSGEEATWEAEDIMREKYPHLF 397
>BP055130
Length = 567
Score = 106 bits (264), Expect(2) = 1e-24
Identities = 55/112 (49%), Positives = 72/112 (64%)
Frame = +2
Query: 284 GNDAIWVIVDRLTKSAHFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRF 343
G I V+VDRL+K HF P ++ +Q+AE ++ IVKLHG+P +IVSDRD FTS F
Sbjct: 179 GFTVIIVVVDRLSKYGHFAPHRANYTSSQVAETFVSTIVKLHGMPRAIVSDRDKAFTSAF 358
Query: 344 WKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHL 395
WK + G+ L +SS+YHPQTDGQ+E + LE LR V E W S+L
Sbjct: 359 WKHFFKLHGTTLNMSSSYHPQTDGQTEALNKCLELYLRCFVHETPRLWVSYL 514
Score = 24.3 bits (51), Expect(2) = 1e-24
Identities = 9/17 (52%), Positives = 13/17 (75%)
Frame = +3
Query: 395 LPLIEFTYNNSYHSSIG 411
LP E+ YN+S+ +SIG
Sbjct: 516 LPWAEYCYNSSFQTSIG 566
>BP054462
Length = 422
Score = 95.1 bits (235), Expect = 3e-20
Identities = 52/141 (36%), Positives = 85/141 (59%), Gaps = 1/141 (0%)
Frame = +1
Query: 403 NNSYHSSIGMAPFEALYGRRCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQS 462
N++Y+ S M+PF+ALYGR + L ++ + E + ++ + SQ
Sbjct: 1 NSNYNRSAKMSPFQALYGREPPVLLQGTTIPSKIAAVNDLQVGRDELLSDLRANLLKSQD 180
Query: 463 RQKTYHDKRRKDLEFQEGDHVFLRVTPMTGVGRALK-SKKLTPKFIGPYQILERVGTVAY 521
+TY +K+R+D+++Q GD VFL++ P A K ++KL+P++ GPY I+ ++G VAY
Sbjct: 181 MMRTYANKKRRDVDYQIGDEVFLKLQPYRRRSLAKKMNEKLSPRYYGPYPIVAKIGAVAY 360
Query: 522 RVGLPPHLSNLHNVFHVSQLQ 542
R+ LP H S +H VFHVS L+
Sbjct: 361 RLELPAH-SRVHPVFHVSLLK 420
>BF177840
Length = 410
Score = 91.3 bits (225), Expect = 4e-19
Identities = 52/130 (40%), Positives = 72/130 (55%)
Frame = +2
Query: 329 SSIVSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQG 388
+SIVSDRD +F S FW++L +G+KL S+ HPQTDGQ+E ++L LLR +
Sbjct: 11 TSIVSDRDTKFISHFWRTLWGKVGTKLLYSTTCHPQTDGQTEVVNKTLSTLLRSVLERNL 190
Query: 389 GTWDSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRIPLCWFESGERVVLGPAIVQQTTE 448
W++ LP IEF YN HS+ +PFE +YG PL +L + E
Sbjct: 191 KMWETWLPHIEFAYNRVVHSTTKHSPFEIVYGYNPLTPLDLLPMPNTYLLKHKDGKAKAE 370
Query: 449 KVQMIQEKIK 458
V+ + EKIK
Sbjct: 371 FVKRLHEKIK 400
>BP085845
Length = 464
Score = 61.2 bits (147), Expect = 5e-10
Identities = 36/106 (33%), Positives = 58/106 (53%)
Frame = -2
Query: 498 KSKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHVSQLQKYVPDPSHVIQSDDV 557
K +K K IGP++ILE V +AY + +L H V HV+ L + + D S I + V
Sbjct: 421 KKRKTESKIIGPFKILEMVCLIAY*LTHSLYLLAAHIVLHVTLLWRNLYDQSQNICHEGV 242
Query: 558 QVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWTGATGESLTWE 603
+ ++ + P+ + D KV+ +R K I V+V+W G +G +WE
Sbjct: 241 XLGEHWSHMEHPIVMVDMKVRCMRPKNIDDVKVIWRGLSG*EKSWE 104
>BP046863
Length = 580
Score = 59.3 bits (142), Expect = 2e-09
Identities = 33/103 (32%), Positives = 63/103 (61%), Gaps = 1/103 (0%)
Frame = +1
Query: 442 IVQQTTEKVQMIQEKIKASQSRQKTYHDKRRKDLEFQEGDHVFLRVTPMTGVGRAL-KSK 500
++++ + ++ + +Q + + +K R+ +++Q G+ VFL++ P A K++
Sbjct: 274 LIEERDALLLELRGNLLKAQDQMRAQANKHRRYVDYQVGNWVFLKLQPYKLQNLAQRKNQ 453
Query: 501 KLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHVSQLQK 543
KL+P+F GP+++LERV VAY + L S +H VFH+S L+K
Sbjct: 454 KLSPRFYGPFKVLERVVQVAY*LDLXSE-SRVHPVFHLSLLEK 579
>TC11573 similar to UP|Q9M6N4 (Q9M6N4) Pol protein integrase region
(Fragment), partial (10%)
Length = 572
Score = 56.6 bits (135), Expect = 1e-08
Identities = 29/90 (32%), Positives = 49/90 (54%)
Frame = +2
Query: 338 RFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPL 397
+FTS+ + + +G ++R SS HPQT+GQ+E + + ++ + E G W LP+
Sbjct: 20 QFTSKQTQDFCDGMGIQMRFSSVKHPQTNGQTEAANKVILKGIKRRLYEAEGRWIDELPI 199
Query: 398 IEFTYNNSYHSSIGMAPFEALYGRRCRIPL 427
+ ++YN SSI PF YG +P+
Sbjct: 200 VLWSYNTMPQSSIKETPF*LTYGADTMLPV 289
>BP064003
Length = 509
Score = 33.1 bits (74), Expect = 0.13
Identities = 31/101 (30%), Positives = 46/101 (44%), Gaps = 6/101 (5%)
Frame = +1
Query: 44 GSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYDFGLNYHP------GKANVVADAL 97
GSRF + +D+ + QK R R W + L G+ + P ++N VADAL
Sbjct: 115 GSRFVIKTDNVATSSFLTQKRAPTRAR-WQDFLSG---GVRHGPRVQAREDQSNKVADAL 282
Query: 98 SRKTLHMSAMMVREFELLEQFRDMSLVCEWSPQSVKLGMLK 138
SRK S LE+ DMS + P+ ++ G+ K
Sbjct: 283 SRKAELAS-------NKLEEIADMSQLKGAIPERIREGLEK 384
>BP048192
Length = 551
Score = 31.6 bits (70), Expect = 0.39
Identities = 13/42 (30%), Positives = 25/42 (58%), Gaps = 3/42 (7%)
Frame = +3
Query: 45 SRFEVFSDHKSLKYLFDQ---KELNMRQRRWLELLKDYDFGL 83
+ F ++D+ ++ Y Q KE+++ + RW +KDYD G+
Sbjct: 192 THFLTYADNNAVGYFIKQGFTKEIHLEKERWQGYIKDYDGGI 317
>TC14209 similar to UP|AAQ72788 (AAQ72788) Hypersensitive-induced response
protein, complete
Length = 1298
Score = 30.8 bits (68), Expect = 0.67
Identities = 16/57 (28%), Positives = 25/57 (43%)
Frame = -1
Query: 13 QLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQ 69
Q V NY THD+EL + H + G++ + F + YL N+R+
Sbjct: 1133 QRNVQNSNYSTHDIELG-------VQLHSIIGAKMQSFKQQRYFCYLLSTIHQNLRK 984
>TC13084
Length = 508
Score = 30.0 bits (66), Expect = 1.1
Identities = 12/24 (50%), Positives = 17/24 (70%)
Frame = +1
Query: 165 DQTEDSDFKFDDQGVLRFRGRICI 188
+ TEDSD+ +++ RFRGR CI
Sbjct: 160 NMTEDSDYDEEEEDYPRFRGRYCI 231
>BP033788
Length = 518
Score = 29.6 bits (65), Expect = 1.5
Identities = 15/43 (34%), Positives = 25/43 (57%)
Frame = +3
Query: 345 KSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQ 387
KSLQ G++++L + P+ D ERT+Q D +I V ++
Sbjct: 75 KSLQTKSGARIQLIPQHLPEGDDSKERTVQVTGDKRQIEVAQE 203
>BG662219
Length = 366
Score = 27.7 bits (60), Expect = 5.7
Identities = 20/57 (35%), Positives = 28/57 (49%), Gaps = 5/57 (8%)
Frame = +3
Query: 465 KTYHDKRRKDLEFQEGDHVFLRVTPM--TGVGRALKSKKLTP---KFIGPYQILERV 516
KT+ RR+ + F+ L VTPM GV R K L K G +++LE+V
Sbjct: 129 KTFRWLRRRPVRFESKSSTPLSVTPMLTPGVARIQKVSSLVSLAMKLQGLWKVLEKV 299
>TC15349 homologue to UP|143C_SOYBN (Q96452) 14-3-3-like protein C (SGF14C),
complete
Length = 1149
Score = 27.3 bits (59), Expect = 7.4
Identities = 14/28 (50%), Positives = 16/28 (57%)
Frame = +1
Query: 582 GKEIPLVRVVWTGATGESLTWELESKML 609
G EI LV VVW +G LTW +ML
Sbjct: 1039 GVEIALVGVVWL*FSGLLLTWIFPKRML 1122
>TC18080 similar to GB|AAL90935.1|19699138|AY090274 AT3g18790/MVE11_17
{Arabidopsis thaliana;}, partial (24%)
Length = 585
Score = 26.9 bits (58), Expect = 9.7
Identities = 20/79 (25%), Positives = 37/79 (46%)
Frame = +3
Query: 130 QSVKLGMLKIDSEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFDDQGVLRFRGRICIP 189
+ VK G + S +R I ++ DV + + R+ E + + F + +P
Sbjct: 30 KGVKSGEVAEVSAAVREILREEEEDVVEEERMRGREMKERLE-----ENEREFVVHVPLP 194
Query: 190 DNEEIKKMILEESHRSSLS 208
D +EI+KM+LE+ L+
Sbjct: 195 DEKEIEKMVLEKKKMELLN 251
>TC18321 similar to UP|ORN_ARATH (Q9ZVE0) Probable oligoribonuclease ,
partial (78%)
Length = 846
Score = 26.9 bits (58), Expect = 9.7
Identities = 14/38 (36%), Positives = 25/38 (64%)
Frame = +2
Query: 422 RCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKA 459
+ R L +ESG +V++ A Q+ KVQ+++EK+K+
Sbjct: 299 KLRSALTGWESGVKVIMQLAG*QKGCLKVQLVKEKLKS 412
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,710,930
Number of Sequences: 28460
Number of extensions: 146581
Number of successful extensions: 683
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 678
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 680
length of query: 617
length of database: 4,897,600
effective HSP length: 96
effective length of query: 521
effective length of database: 2,165,440
effective search space: 1128194240
effective search space used: 1128194240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC138199.5


