

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138130.16 + phase: 0
(425 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
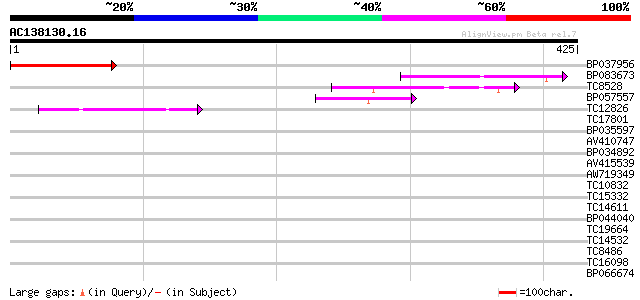
Score E
Sequences producing significant alignments: (bits) Value
BP037956 146 6e-36
BP083673 57 8e-09
TC8528 weakly similar to GB|AAO42978.1|28416895|BT004732 At3g139... 54 4e-08
BP057557 48 3e-06
TC12826 weakly similar to UP|Q9LVG7 (Q9LVG7) Ankyrin-like protei... 44 4e-05
TC17801 weakly similar to GB|AAO42978.1|28416895|BT004732 At3g13... 40 0.001
BP035597 39 0.002
AV410747 36 0.014
BP034892 30 0.58
AV415539 30 0.76
AW719349 30 0.76
TC10832 weakly similar to UP|Q9SEE9 (Q9SEE9) Arginine/serine-ric... 30 0.99
TC15332 similar to UP|PPA1_LYCES (P27061) Acid phosphatase precu... 29 1.3
TC14611 similar to UP|Q8LPB2 (Q8LPB2) Glycine-rich RNA binding p... 29 1.7
BP044040 29 1.7
TC19664 similar to UP|Q9ZSQ7 (Q9ZSQ7) Protein phosphatase 2C hom... 28 2.2
TC14532 similar to UP|Q9FYA8 (Q9FYA8) SC35-like splicing factor ... 28 2.2
TC8486 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {Ara... 28 2.2
TC16098 homologue to UP|Q8LAQ5 (Q8LAQ5) Flower pigmentation prot... 28 2.9
BP066674 27 6.4
>BP037956
Length = 476
Score = 146 bits (369), Expect = 6e-36
Identities = 68/80 (85%), Positives = 74/80 (92%)
Frame = +3
Query: 1 MNGKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDR 60
MNGKVSIL+DFVS AASFHYLTREEET+FHLAVRYGCYDA VFLVQVSNGTNLLHCQDR
Sbjct: 183 MNGKVSILEDFVSSCAASFHYLTREEETIFHLAVRYGCYDAFVFLVQVSNGTNLLHCQDR 362
Query: 61 YGNSVLHLAVSGGRHKMTDF 80
+GNS LHLAV GGRHK+ ++
Sbjct: 363 FGNSALHLAVIGGRHKVRNY 422
>BP083673
Length = 437
Score = 56.6 bits (135), Expect = 8e-09
Identities = 34/132 (25%), Positives = 68/132 (50%), Gaps = 7/132 (5%)
Frame = -1
Query: 294 NNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVILPHNQ 353
++IALF SL+VV+V S+V + + ++ I +K+MW+A ++ ++A ++V++ +
Sbjct: 434 DSIALFISLAVVVVQTSVVVIESQAKKQMMAIINKLMWLACVLVSVAFLALSFVVVGKEE 255
Query: 354 GMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKRKEIGDG-------AAESD 406
+WL++ + +G + T + ++ H + S R RK + A SD
Sbjct: 254 --KWLAIGVTIIGTTIMATTLGTMCYWVIRHRIEASNLRNIRKSSLESKSRSFSQPALSD 81
Query: 407 KESENSDFQSSY 418
E N +F+ Y
Sbjct: 80 SELLNHEFKKMY 45
>TC8528 weakly similar to GB|AAO42978.1|28416895|BT004732 At3g13950
{Arabidopsis thaliana;}, partial (17%)
Length = 788
Score = 54.3 bits (129), Expect = 4e-08
Identities = 43/162 (26%), Positives = 75/162 (45%), Gaps = 21/162 (12%)
Frame = +2
Query: 242 NARNTIVLVAVLIATVTFAAGISPPGGVYQ-------------------EGPMRGKSMVG 282
N R ++ L A +IAT+ F +PPGGV+Q + P V
Sbjct: 146 NMRGSVSLTASIIATMAFQLATNPPGGVFQANGGDSVAKIKSCLDNDAIQCPGEAVLAVV 325
Query: 283 RTSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYV 342
+ +F N I+ +SLSV ++LVS +P + + +L I M +++ +A Y+
Sbjct: 326 NEDDYSLFLTFNTISFISSLSVCLLLVSGIPLKHRVIIWILSIG---MCISITSLALTYL 496
Query: 343 AATWVILPHNQGMQWLSVLLLAL--GGGSLGTIFIGLSVMLV 382
A ++ P++ W +V LAL G LG + + L + ++
Sbjct: 497 VAASMVTPNH---VWGNVFALALIIWIGLLGIVSVYLIIRMI 613
>BP057557
Length = 497
Score = 48.1 bits (113), Expect = 3e-06
Identities = 29/79 (36%), Positives = 45/79 (56%), Gaps = 3/79 (3%)
Frame = -1
Query: 230 KHYHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPG---GVYQEGPMRGKSMVGRTSA 286
K ++H L NA N+ +VAVLIATV FAA + PG ++ G + + +A
Sbjct: 407 KKLKKLHISGLNNAINSATVVAVLIATVAFAAIFTVPGQYVEAKEQSFSLGPANIANNAA 228
Query: 287 FKVFAISNNIALFTSLSVV 305
F +F + + +ALF SL+V+
Sbjct: 227 FMIFFVFDILALFISLAVI 171
>TC12826 weakly similar to UP|Q9LVG7 (Q9LVG7) Ankyrin-like protein, partial
(23%)
Length = 528
Score = 44.3 bits (103), Expect = 4e-05
Identities = 34/123 (27%), Positives = 64/123 (51%)
Frame = +2
Query: 22 LTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYGNSVLHLAVSGGRHKMTDFL 81
+ ++ +T H+AV+ + + LV ++ + ++ D GN+ LH+A R K+ L
Sbjct: 29 IDKKGQTALHMAVKGQNLELVDELVTLTPSS--VNMVDAKGNTALHIATRKSRLKIIQRL 202
Query: 82 INKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFIRAGGKRSIQSSSFSLELDKNN 141
++ +++D N N G TALDI +++ + LQ A +SI+SSS + L+
Sbjct: 203 LDCSEIDTNVINKSGETALDIAERSSHLEITNSLQD--HGAQNAKSIRSSSRNPALELKR 376
Query: 142 SPS 144
+ S
Sbjct: 377 TVS 385
>TC17801 weakly similar to GB|AAO42978.1|28416895|BT004732 At3g13950
{Arabidopsis thaliana;}, partial (22%)
Length = 423
Score = 39.7 bits (91), Expect = 0.001
Identities = 26/91 (28%), Positives = 44/91 (47%), Gaps = 19/91 (20%)
Frame = +2
Query: 244 RNTIVLVAVLIATVTFAAGISPPGGVYQEG-----------------PMRGKSMVG--RT 284
R + + A +IAT+TF +PPGGV+Q G++++G
Sbjct: 149 RGGLSMTASIIATMTFQLATNPPGGVFQANGGVPKTDARICWHNNTIQCPGEAVLGVVYR 328
Query: 285 SAFKVFAISNNIALFTSLSVVIVLVSIVPFR 315
+ F I N ++ +SLSV ++LVS +P +
Sbjct: 329 HTYTNFLICNTLSFISSLSVCLLLVSGIPLK 421
>BP035597
Length = 467
Score = 38.5 bits (88), Expect = 0.002
Identities = 23/69 (33%), Positives = 36/69 (51%)
Frame = -1
Query: 28 TVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYGNSVLHLAVSGGRHKMTDFLINKTKL 87
T H A G D ++ LV+ L C+D G+ LH+AV GG + L+++ +
Sbjct: 461 TALHYAACKGHKDVVLMLVEAGWD---LECEDEEGHVPLHMAVEGGDFETVPILVDQ-GV 294
Query: 88 DINTRNSEG 96
++N RNS G
Sbjct: 293 NLNARNSXG 267
>AV410747
Length = 312
Score = 35.8 bits (81), Expect = 0.014
Identities = 15/30 (50%), Positives = 22/30 (73%)
Frame = +3
Query: 242 NARNTIVLVAVLIATVTFAAGISPPGGVYQ 271
N R ++ L+A +IAT+TF +PPGGV+Q
Sbjct: 150 NMRGSLSLMASIIATMTFQLATNPPGGVFQ 239
>BP034892
Length = 483
Score = 30.4 bits (67), Expect = 0.58
Identities = 18/54 (33%), Positives = 27/54 (49%), Gaps = 1/54 (1%)
Frame = +1
Query: 42 LVFLVQVSNGTNLLHCQDRYGNSVL-HLAVSGGRHKMTDFLINKTKLDINTRNS 94
L++ V VS G L HC YG L H+ +SGG+H + + K + +S
Sbjct: 25 LLWSVLVSVGV-LFHCDGAYGEKNLPHMTISGGKHHVAIQRLQSFKASLTRHDS 183
>AV415539
Length = 419
Score = 30.0 bits (66), Expect = 0.76
Identities = 18/60 (30%), Positives = 27/60 (45%), Gaps = 4/60 (6%)
Frame = +2
Query: 142 SPSPAYRLSPSRRY----IPNEMEVLTEMVSYDCISPPPVSKSSDSRSPQPQASERFENG 197
SPSP+ SPSR P+ + + C S PP+ S P P+++ R +G
Sbjct: 65 SPSPSSSTSPSRSSSPAAAPSHSATSRQSTASPCPSSPPLFSPEHSPPPPPRSATRAGSG 244
>AW719349
Length = 143
Score = 30.0 bits (66), Expect = 0.76
Identities = 12/40 (30%), Positives = 21/40 (52%), Gaps = 2/40 (5%)
Frame = -3
Query: 199 YNPYYVSPTNLVKQKHH--HNKGKIENVNHTKRKHYHEMH 236
+ PY+ S L+ HH H+ +++H+ R H+H H
Sbjct: 123 FPPYFHSHHTLLHSLHHSHHSSSYFHSLHHSLRNHHHSHH 4
>TC10832 weakly similar to UP|Q9SEE9 (Q9SEE9) Arginine/serine-rich protein,
partial (30%)
Length = 539
Score = 29.6 bits (65), Expect = 0.99
Identities = 23/66 (34%), Positives = 30/66 (44%)
Frame = +3
Query: 124 GKRSIQSSSFSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSSDS 183
G+RS S S S +++SPS + S SR L+ S+ S P S S
Sbjct: 120 GRRSPPSGSVSGSSSRSSSPSSSRSSSRSRS--------LSRSRSFSSSSSSPSRGGSRS 275
Query: 184 RSPQPQ 189
RSP PQ
Sbjct: 276 RSPPPQ 293
>TC15332 similar to UP|PPA1_LYCES (P27061) Acid phosphatase precursor 1 ,
partial (89%)
Length = 1040
Score = 29.3 bits (64), Expect = 1.3
Identities = 17/52 (32%), Positives = 23/52 (43%)
Frame = -1
Query: 139 KNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSSDSRSPQPQA 190
+ +PSP+ S +RR P + C PPP S S SP P+A
Sbjct: 353 RKRNPSPSCTPSRARRIPPAPSSTAS-----GCSPPPPASSRRTSASPPPRA 213
>TC14611 similar to UP|Q8LPB2 (Q8LPB2) Glycine-rich RNA binding protein,
partial (20%)
Length = 760
Score = 28.9 bits (63), Expect = 1.7
Identities = 10/32 (31%), Positives = 18/32 (56%)
Frame = -2
Query: 214 HHHNKGKIENVNHTKRKHYHEMHQEALLNARN 245
HHH++ ++++ HYH H +AL A +
Sbjct: 333 HHHHQSHHLHISYEHSPHYHHHHHQALEGAHD 238
>BP044040
Length = 491
Score = 28.9 bits (63), Expect = 1.7
Identities = 25/98 (25%), Positives = 40/98 (40%)
Frame = +3
Query: 127 SIQSSSFSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSSDSRSP 186
S SSS S ++ N +P+P RL+P++ PN P S+ + P
Sbjct: 159 SSSSSSSSAPINPNRNPTP--RLTPTQNPNPNS---------------KPRPASTTNHEP 287
Query: 187 QPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIENV 224
PQ + R ++P+ +LV + G NV
Sbjct: 288 SPQKTNRNPGDGFDPF----DSLVTSSNRSQNGYSLNV 389
>TC19664 similar to UP|Q9ZSQ7 (Q9ZSQ7) Protein phosphatase 2C homolog,
partial (9%)
Length = 426
Score = 28.5 bits (62), Expect = 2.2
Identities = 17/71 (23%), Positives = 26/71 (35%), Gaps = 1/71 (1%)
Frame = -2
Query: 149 LSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSSDSRSPQPQASERFENGT-YNPYYVSPT 207
L P + ++P + SPPP P+ S +N Y+P+
Sbjct: 353 LVPQQPFLPPRTHIRAPFAVVRGSSPPPAGAQVQVH*PR*HPSFHHQNNPRYSPFTSKHV 174
Query: 208 NLVKQKHHHNK 218
N +K H H K
Sbjct: 173 NQIKPNHKHAK 141
>TC14532 similar to UP|Q9FYA8 (Q9FYA8) SC35-like splicing factor SCL30a, 30a
kD, partial (71%)
Length = 1073
Score = 28.5 bits (62), Expect = 2.2
Identities = 27/98 (27%), Positives = 36/98 (36%), Gaps = 9/98 (9%)
Frame = +3
Query: 113 RQLQAIFIRAGGKRSIQ----SSSFSLELDKNNSP-----SPAYRLSPSRRYIPNEMEVL 163
R+L +F K+ + S D+ SP SP Y SP R+
Sbjct: 375 RELTVVFAEENRKKPTEMRHRERSRGRSYDRRRSPPRYSRSPRYSRSPPPRHRSRSR--- 545
Query: 164 TEMVSYDCISPPPVSKSSDSRSPQPQASERFENGTYNP 201
S D SPPP + DSRS P+ +Y P
Sbjct: 546 ----SRDYYSPPPKRR--DSRSVSPEERRYSRERSYTP 641
>TC8486 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {Arabidopsis
thaliana;}, partial (25%)
Length = 856
Score = 28.5 bits (62), Expect = 2.2
Identities = 15/44 (34%), Positives = 22/44 (49%)
Frame = -1
Query: 180 SSDSRSPQPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIEN 223
SSD+ SPQP +E + +Y PT QK H G +++
Sbjct: 754 SSDNHSPQPATAETASESRWG*HY--PTR*PDQKRVHGAGTLDS 629
>TC16098 homologue to UP|Q8LAQ5 (Q8LAQ5) Flower pigmentation protein ATAN11,
partial (79%)
Length = 919
Score = 28.1 bits (61), Expect = 2.9
Identities = 16/43 (37%), Positives = 22/43 (50%), Gaps = 2/43 (4%)
Frame = +1
Query: 148 RLSPSRRYI--PNEMEVLTEMVSYDCISPPPVSKSSDSRSPQP 188
R+SPS + P LT+ V+ SPPP + S+ SP P
Sbjct: 280 RISPSNTHTHPPRPFSSLTKSVNALTSSPPPATSSACGGSPTP 408
>BP066674
Length = 181
Score = 26.9 bits (58), Expect = 6.4
Identities = 9/19 (47%), Positives = 12/19 (62%)
Frame = +3
Query: 201 PYYVSPTNLVKQKHHHNKG 219
PY + P L+K HH+N G
Sbjct: 54 PYLLEPPKLIKXAHHNNXG 110
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,409,705
Number of Sequences: 28460
Number of extensions: 111233
Number of successful extensions: 890
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 860
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 877
length of query: 425
length of database: 4,897,600
effective HSP length: 93
effective length of query: 332
effective length of database: 2,250,820
effective search space: 747272240
effective search space used: 747272240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC138130.16


