

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138130.14 + phase: 0
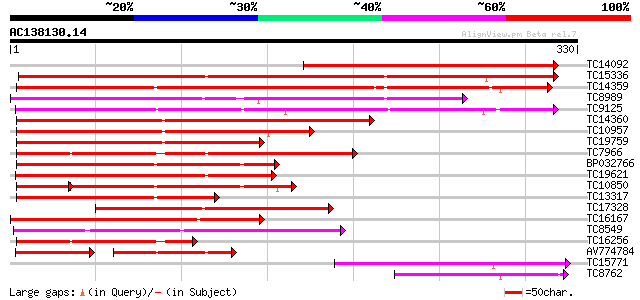
(330 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14092 weakly similar to UP|Q94KE6 (Q94KE6) Dihydro-flavanoid r... 274 2e-74
TC15336 255 6e-69
TC14359 similar to UP|AAQ88099 (AAQ88099) NADPH-dependent cinnam... 239 6e-64
TC8989 weakly similar to UP|Q9FGH3 (Q9FGH3) Dihydroflavonol 4-re... 204 1e-53
TC9125 similar to UP|Q40316 (Q40316) Vestitone reductase, complete 203 3e-53
TC14360 similar to UP|O04391 (O04391) Cinnamyl alcohol dehydroge... 195 8e-51
TC10957 similar to UP|Q8L9G4 (Q8L9G4) Cinnamyl-alcohol dehydroge... 182 9e-47
TC19759 similar to UP|Q8L9G4 (Q8L9G4) Cinnamyl-alcohol dehydroge... 162 9e-41
TC7966 similar to UP|Q84V25 (Q84V25) Quinone oxidoreductase, par... 157 2e-39
BP032766 138 1e-33
TC19621 similar to UP|Q8VZH7 (Q8VZH7) Cinnamoyl-CoA reductase ,... 137 2e-33
TC10850 homologue to UP|Q9S6Y4 (Q9S6Y4) Dihydroflavanol reductas... 100 5e-30
TC13317 similar to UP|Q9SPJ5 (Q9SPJ5) Dihydroflavonol-4-reductas... 115 8e-27
TC17328 weakly similar to UP|Q9FGH3 (Q9FGH3) Dihydroflavonol 4-r... 115 8e-27
TC16167 similar to UP|Q9FGH3 (Q9FGH3) Dihydroflavonol 4-reductas... 115 1e-26
TC8549 weakly similar to UP|Q8LEH3 (Q8LEH3) Cinnamoyl-CoA reduct... 103 3e-23
TC16256 similar to UP|Q7Y0H8 (Q7Y0H8) Cinnamoyl CoA reductase, p... 96 8e-21
AV774784 57 7e-16
TC15771 similar to UP|AAR27015 (AAR27015) Dihydroflavonal-4-redu... 76 9e-15
TC8762 similar to UP|O65880 (O65880) Cinnamoyl CoA reductase , ... 73 6e-14
>TC14092 weakly similar to UP|Q94KE6 (Q94KE6) Dihydro-flavanoid
reductase-like protein, partial (32%)
Length = 721
Score = 274 bits (700), Expect = 2e-74
Identities = 126/148 (85%), Positives = 135/148 (91%)
Frame = +1
Query: 172 WDYCKENGIDLVTILPSFIIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDV 231
W+YCKENGI LVT+LPSFIIGP+LP DLCSTASDVLGL KGETEKFQWHGRMGYVHIDDV
Sbjct: 1 WEYCKENGIHLVTVLPSFIIGPSLPPDLCSTASDVLGLLKGETEKFQWHGRMGYVHIDDV 180
Query: 232 ALCHILLYENKASDGRYLCSSKIMDNDDLVGMLANRYPGFPIPKRFKKLDRPHYELNTGK 291
ALCHILL+EN+AS GRYLCSS +MDNDDLV +LA RYP PIPKRF+KLDRPHY+LN GK
Sbjct: 181 ALCHILLFENEASHGRYLCSSTVMDNDDLVALLATRYPSLPIPKRFQKLDRPHYDLNIGK 360
Query: 292 LESLGFKFKSVEEMFDDCFASFVEQGHL 319
L SLGFKFKSVEEMFDDC AS VEQGHL
Sbjct: 361 LSSLGFKFKSVEEMFDDCIASLVEQGHL 444
>TC15336
Length = 1145
Score = 255 bits (652), Expect = 6e-69
Identities = 138/317 (43%), Positives = 201/317 (62%), Gaps = 3/317 (0%)
Frame = +2
Query: 6 CVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLMEE 65
CVTG +GF+A++L++ LL G+ V TVR+ G +KV L +L GA ERLK+++ADL+ E
Sbjct: 68 CVTGGTGFIAAYLVQALLEKGHTVRTTVRNPGDVEKVGFLMELSGAKERLKILKADLLNE 247
Query: 66 NSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLAS 125
SFD A+ G GVFH ASPVL ++ +A +++P ++G LNVL SC K + RVVL S
Sbjct: 248 GSFDEAVSGVDGVFHTASPVLVPYDDNIQATLIDPCIKGALNVLNSCVK-ANVKRVVLTS 424
Query: 126 SSSAVRVRADFDPNIPIDESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCKENGIDLVTI 185
S S++R R D P++ES WS E C++ WY +KTLAEK AW KE+G+DLV +
Sbjct: 425 SCSSIRYRDDEQQVSPLNESHWSDPEYCKRYNLWYAYAKTLAEKEAWKIAKESGMDLVVV 604
Query: 186 LPSFIIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDVALCHILLYENKASD 245
PSF++GP L ST +L + KG ++ + +G+VHI DV H+L E +
Sbjct: 605 NPSFVVGPLLAPQPTSTLLMILSIIKGLKGEYP-NTTVGFVHIHDVVGAHLLAMEEPKAS 781
Query: 246 GRYLCSSKIMDNDDLVGMLANRYPGFPIPKR--FKKLDRPHYELNTGKLESLGF-KFKSV 302
GR +CSS + ++ ML ++YP +P K+ ++ D + ++T K+ +LGF FKS+
Sbjct: 782 GRLVCSSTVAHWSQIIEMLQSQYPFYPYEKKCGSQEGDNNTHSMDTTKITALGFPAFKSL 961
Query: 303 EEMFDDCFASFVEQGHL 319
E+MFDDC SF E+G L
Sbjct: 962 EQMFDDCIKSFQEKGFL 1012
>TC14359 similar to UP|AAQ88099 (AAQ88099) NADPH-dependent cinnamyl alcohol
dehydrogenase, partial (98%)
Length = 1403
Score = 239 bits (609), Expect = 6e-64
Identities = 133/316 (42%), Positives = 199/316 (62%), Gaps = 4/316 (1%)
Frame = +1
Query: 5 VCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLME 64
VCVTGASG++ASW++K LL GY V TVRD +KV+HL L+GA ERL L +A+L+E
Sbjct: 73 VCVTGASGYIASWIVKFLLHRGYTVKATVRDPNDSKKVDHLLNLDGAKERLHLFKANLLE 252
Query: 65 ENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLA 124
E SFD+ + GC GVFH ASP H DP+ E+L+PAV+GTLNVL+SC +P L RVVL
Sbjct: 253 EGSFDSVVQGCHGVFHTASP-FYHDVKDPQVELLDPAVKGTLNVLKSCVNSPTLKRVVLT 429
Query: 125 SSSSAVRVRA-DFDPNIPIDESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCKENGIDLV 183
SS +AV P++ +DE+ ++ L + + WY +SKTLAE+AAW + +EN ID+V
Sbjct: 430 SSIAAVAYNGKPRTPDVVVDETWFTDPVLNREAKMWYVLSKTLAEEAAWKFVRENNIDMV 609
Query: 184 TILPSFIIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDVALCHILLYENKA 243
TI P+ +IGP L L ++A+ VL G + F + G+V++ DVA HI YE +
Sbjct: 610 TINPAMVIGPLLQPVLNTSAASVLNFVNG-AQTFA-NASFGWVNVKDVANAHIQAYEIAS 783
Query: 244 SDGRYLCSSKIMDNDDLVGMLANRYPGFPIPKRFKKLDRPH---YELNTGKLESLGFKFK 300
+ GR+ +++ + ++V +L YP +P++ D+P+ Y+++ K +SLG ++
Sbjct: 784 ASGRHCLVERVVHHSEIVKILRELYPTLQLPEKCAD-DKPYVPTYQVSKEKAKSLGIEYI 960
Query: 301 SVEEMFDDCFASFVEQ 316
+E + S E+
Sbjct: 961 PLEVSLKETVESLKEK 1008
>TC8989 weakly similar to UP|Q9FGH3 (Q9FGH3) Dihydroflavonol
4-reductase-like (Cinnamoyl-CoA reductase-like protein),
partial (77%)
Length = 880
Score = 204 bits (520), Expect = 1e-53
Identities = 112/270 (41%), Positives = 162/270 (59%), Gaps = 4/270 (1%)
Frame = +2
Query: 1 MEHKVCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQA 60
M +CVTGASG + SW++K LL GY V TV+DL +++ +HL +EGA RL+L +
Sbjct: 89 MSKVLCVTGASGAIGSWVVKLLLDRGYTVHATVQDLKDEKETKHLEAMEGAKGRLRLFEM 268
Query: 61 DLMEENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVR 120
DL++ NS + GC GV H+A P + DP+ +ILEPA++GT+NVL++ K R
Sbjct: 269 DLLDTNSIATTVKGCNGVIHLACPNVIGEVTDPEKQILEPAIKGTVNVLKAA-KEAGAER 445
Query: 121 VVLASSSSAVRVRADFDPNIPID----ESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCK 176
VV SS SA+ PN P D E W+ LE C++ +YP++KTLAEKA W++ K
Sbjct: 446 VVATSSISAITP----SPNWPADKIKGEDCWTDLEYCKEKGLYYPIAKTLAEKAGWEFAK 613
Query: 177 ENGIDLVTILPSFIIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDVALCHI 236
E G D+V I P +G + + S+ + + G+ KG+ E ++ MG H D+AL HI
Sbjct: 614 ETGFDVVMINPGTALGSLISPRINSSMAVLAGVLKGDKETYE-DFFMGMAHFKDIALAHI 790
Query: 237 LLYENKASDGRYLCSSKIMDNDDLVGMLAN 266
L +ENK + GR+LC I D V M+A+
Sbjct: 791 LAFENKKAAGRHLCVEAIRHYSDFVAMVAD 880
>TC9125 similar to UP|Q40316 (Q40316) Vestitone reductase, complete
Length = 1200
Score = 203 bits (517), Expect = 3e-53
Identities = 126/327 (38%), Positives = 193/327 (58%), Gaps = 11/327 (3%)
Frame = +3
Query: 4 KVCVTGASGFLASWLIKRLLLSGYHVIGTVR-DLGKKQKVEHLWKLEGAT-ERLKLVQAD 61
+VCVTG +GFLASW+IKRLL GY V T+R + G K+ V L L GAT E+L++ AD
Sbjct: 57 RVCVTGGTGFLASWIIKRLLEDGYSVNTTIRSNSGGKRDVSFLTNLPGATSEKLQIFNAD 236
Query: 62 LMEENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRV 121
L SF A+ GC G+FH A+PV + N+P+ + + V G L +L++C + + RV
Sbjct: 237 LCIPESFGPAVEGCVGIFHTATPV-DFAVNEPEEVVTKRTVDGALGILKACVNSKTVKRV 413
Query: 122 VLASSSSAVRVRADFDPNIPIDESSWSSLELCEKLQAW---YPMSKTLAEKAAWDYCKEN 178
V SS SA D +DES WS ++L K++ + Y +SKTLAEKA ++ +++
Sbjct: 414 VYTSSGSAASFSGK-DGGDALDESYWSDVDLLRKVKPFGWSYAVSKTLAEKAVLEFGEKH 590
Query: 179 GIDLVTILPSFIIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDVALCHILL 238
G+++VT++P+F++GP + L + L L G+ E+ R VH+DDVA HI L
Sbjct: 591 GLEVVTLIPTFVVGPWVCPKLPDSVERALVLVLGKKEQIGVI-RFHMVHVDDVARAHIFL 767
Query: 239 YENKASDGRYLCSSKIMDNDDLVGMLANRYPGFPIP-----KRFKKLDRPHYELNTGKLE 293
E+ GRY CS I+ +++ +L+ +YP + IP K + +P +L + K+
Sbjct: 768 LEHPNPKGRYNCSPFIVPIEEMSKLLSAKYPEYQIPSVEELKGIEGFKQP--DLISNKIR 941
Query: 294 SLGFKFK-SVEEMFDDCFASFVEQGHL 319
GF+FK S+E++FDD E+G+L
Sbjct: 942 DAGFEFKYSLEDLFDDAIQCCKEKGYL 1022
>TC14360 similar to UP|O04391 (O04391) Cinnamyl alcohol dehydrogenase ,
partial (68%)
Length = 821
Score = 195 bits (496), Expect = 8e-51
Identities = 104/209 (49%), Positives = 141/209 (66%), Gaps = 1/209 (0%)
Frame = +3
Query: 5 VCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLME 64
VCVTGASG++ASW++K LL G V TVRD +KV HL +L+GA ERL L +A+L+E
Sbjct: 78 VCVTGASGYIASWIVKFLLQRGNTVKATVRDPNDPKKVGHLLRLDGARERLHLFKANLLE 257
Query: 65 ENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLA 124
E SF++ + GC GVFH ASP + DP+AE+L+PAV+GTLNVL+SC +P+L RVVL
Sbjct: 258 EGSFNSVVQGCHGVFHTASPFYYDV-EDPQAELLDPAVKGTLNVLKSCANSPSLKRVVLT 434
Query: 125 SSSSAVRVRA-DFDPNIPIDESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCKENGIDLV 183
SS +AV P++ +DE+ +S + WY +SKTLAE AAW + +EN ID+V
Sbjct: 435 SSIAAVAYNGKSRTPDVVVDETWFSDPHFNLEANMWYVLSKTLAEDAAWKFARENNIDMV 614
Query: 184 TILPSFIIGPNLPTDLCSTASDVLGLFKG 212
TI P+ +IGP L L ++A+ VL G
Sbjct: 615 TINPAMVIGPLLQPVLNTSAAAVLNFVNG 701
>TC10957 similar to UP|Q8L9G4 (Q8L9G4) Cinnamyl-alcohol dehydrogenase-like
protein, partial (52%)
Length = 765
Score = 182 bits (461), Expect = 9e-47
Identities = 97/177 (54%), Positives = 125/177 (69%), Gaps = 4/177 (2%)
Frame = +1
Query: 5 VCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLME 64
VCVTG SG++ASW++K LL +GY V TVRD G KVEHL KL+GA ERL+L +ADL+E
Sbjct: 238 VCVTGGSGYIASWIVKFLLENGYTVRATVRDPGNHYKVEHLVKLDGAKERLQLFKADLLE 417
Query: 65 ENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLA 124
E SFD+AI GC GVFHIASPV + DP+AE+++PAV+GTLNVL+SC K+P++ RVVL
Sbjct: 418 EGSFDSAIEGCNGVFHIASPVRMFV-EDPQAELIDPAVKGTLNVLKSCAKSPSVKRVVLT 594
Query: 125 SSSSAVRVRA-DFDPNIPIDESSWSS---LELCEKLQAWYPMSKTLAEKAAWDYCKE 177
SS+SAV P + +DE+ +S L + WY +SKTLAE AW + E
Sbjct: 595 SSTSAVLFNGRPKSPEVVVDETWFSDPDFLRESKLYHLWYTISKTLAEDTAWKFANE 765
>TC19759 similar to UP|Q8L9G4 (Q8L9G4) Cinnamyl-alcohol dehydrogenase-like
protein, partial (46%)
Length = 468
Score = 162 bits (409), Expect = 9e-41
Identities = 84/145 (57%), Positives = 109/145 (74%), Gaps = 1/145 (0%)
Frame = +3
Query: 5 VCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLME 64
VCVTGASG++ASW++K LL GY V TVRDL KVEHL KL+ A ERL+L +ADL+E
Sbjct: 18 VCVTGASGYIASWIVKFLLEHGYTVRATVRDLSNPNKVEHLVKLDVAKERLQLFKADLLE 197
Query: 65 ENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLA 124
E SFD+ I GC GVFH+ASPVL + DP+AE+++PAV+GTLNVL+SC K+P++ RVVL
Sbjct: 198 EGSFDSVIQGCHGVFHVASPVLMFV-EDPQAELIDPAVKGTLNVLKSCAKSPSVKRVVLT 374
Query: 125 SSSSAVRVRA-DFDPNIPIDESSWS 148
SS+SAV P + +DE+ +S
Sbjct: 375 SSTSAVLFNGRPKSPEVVVDETWFS 449
>TC7966 similar to UP|Q84V25 (Q84V25) Quinone oxidoreductase, partial (98%)
Length = 1908
Score = 157 bits (398), Expect = 2e-39
Identities = 92/199 (46%), Positives = 121/199 (60%), Gaps = 1/199 (0%)
Frame = -1
Query: 5 VCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLME 64
+CVTGA GF+ASW++K LL GY V GTVR+ K HL +LEGA++RL L++ DL+E
Sbjct: 588 ICVTGAGGFIASWMVKLLLQKGYTVRGTVRN-PDDPKNGHLRELEGASDRLTLIKVDLLE 412
Query: 65 ENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLA 124
NS A+ G GVFH ASPV D E++EPAV G NV+ + + + RVV
Sbjct: 411 LNSVRAAVHGSHGVFHTASPV-----TDNPEEMVEPAVNGAKNVIIAAAE-AKVRRVVFT 250
Query: 125 SSSSAVRVRADFDPNIPIDESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCKENGIDLVT 184
SS AV + + +DES WS LE C+ + WY K +AE+AAWD KE G+DLV
Sbjct: 249 SSIGAVYMDPKRSVDSVVDESCWSDLEFCKDTKNWYCYGKAVAEQAAWDTAKEKGVDLVV 70
Query: 185 ILPSFIIGPNL-PTDLCST 202
+ P ++GP L PT ST
Sbjct: 69 VNPVLVLGPLLQPTINAST 13
>BP032766
Length = 535
Score = 138 bits (347), Expect = 1e-33
Identities = 70/153 (45%), Positives = 100/153 (64%)
Frame = +1
Query: 5 VCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLME 64
VCVTGA+GF+ SWL+ RL+ GY V TVRD +KV+HL +L A +L L +ADL E
Sbjct: 76 VCVTGAAGFIGSWLVMRLIERGYTVRATVRDPANMKKVKHLLELPDAKTKLSLWKADLAE 255
Query: 65 ENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLA 124
E SFD AI GC GVFH+A+P ++ S DP+ E+++P + G L++L++C K + R+V
Sbjct: 256 EGSFDEAIRGCTGVFHVATP-MDFESKDPENEVIKPTINGLLDILKACEKAKTVRRLVFT 432
Query: 125 SSSSAVRVRADFDPNIPIDESSWSSLELCEKLQ 157
SS+ V V P IDE+ WS +E C +++
Sbjct: 433 SSAGTVDVTE--HPKPVIDETCWSDIEFCLRVK 525
>TC19621 similar to UP|Q8VZH7 (Q8VZH7) Cinnamoyl-CoA reductase , partial
(20%)
Length = 485
Score = 137 bits (345), Expect = 2e-33
Identities = 73/152 (48%), Positives = 98/152 (64%)
Frame = +3
Query: 4 KVCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLM 63
KVCVTGA GF+ASWL+K LL GYH + D K EHL KLE A+E L L +ADL+
Sbjct: 30 KVCVTGAGGFVASWLVKLLLSKGYHCSWXLSDXPGSPKYEHLLKLEKASENLTLFKADLL 209
Query: 64 EENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVL 123
S +AI+GC VFH+ASPV + S++P+ E++EPAV+GT NVL + + + RVV
Sbjct: 210 NYESVHSAILGCTAVFHVASPVPSTTSSNPEVEVIEPAVKGTANVLEASLQ-AKVERVVF 386
Query: 124 ASSSSAVRVRADFDPNIPIDESSWSSLELCEK 155
SS +AV + + + IDES WS + C+K
Sbjct: 387 VSSEAAVCMSPNLPKDKVIDESYWSDKDYCKK 482
>TC10850 homologue to UP|Q9S6Y4 (Q9S6Y4) Dihydroflavanol reductase 3,
partial (51%)
Length = 594
Score = 100 bits (249), Expect(2) = 5e-30
Identities = 56/135 (41%), Positives = 83/135 (61%), Gaps = 3/135 (2%)
Frame = +2
Query: 36 LGKKQKVEHLWKLEGATERLKLVQADLMEENSFDNAIMGCKGVFHIASPVLNHISNDPKA 95
L +KV+HL +L A +L L +ADL EE SFD AI GC GVFH+A+P ++ S DP+
Sbjct: 197 LANMKKVKHLLELPEAKTKLTLWKADLAEEGSFDEAIKGCTGVFHVATP-MDFESKDPEN 373
Query: 96 EILEPAVQGTLNVLRSCRKNPALVRVVLASSSSAVRVRADFDPNIPIDESSWSSLELCE- 154
E+++P + G L+++++C+K + R+V SS+ + V DES WS +E C
Sbjct: 374 EVIKPTINGVLDIMKACQKAKTVRRLVFTSSAGTLNVIE--HQKQMFDESCWSDVEFCRR 547
Query: 155 -KLQAW-YPMSKTLA 167
K+ W Y +SKTLA
Sbjct: 548 VKMTGWMYFVSKTLA 592
Score = 47.0 bits (110), Expect(2) = 5e-30
Identities = 21/33 (63%), Positives = 25/33 (75%)
Frame = +1
Query: 5 VCVTGASGFLASWLIKRLLLSGYHVIGTVRDLG 37
VCVTGA+GF+ SWL+ RL+ GY V TVRD G
Sbjct: 103 VCVTGAAGFIGSWLVMRLMERGYMVRATVRDPG 201
>TC13317 similar to UP|Q9SPJ5 (Q9SPJ5) Dihydroflavonol-4-reductase DFR1,
partial (36%)
Length = 426
Score = 115 bits (289), Expect = 8e-27
Identities = 58/119 (48%), Positives = 82/119 (68%), Gaps = 1/119 (0%)
Frame = +2
Query: 5 VCVTGASGFLASWLIKRLLLSGYHVIGTV-RDLGKKQKVEHLWKLEGATERLKLVQADLM 63
VCVTG++GF+ SWL+ RL+ GY V TV RD +KV+HL +L GA L + ADL
Sbjct: 68 VCVTGSTGFIGSWLVMRLMERGYMVRATVQRDPDNMKKVKHLLELPGAKTNLTIWNADLT 247
Query: 64 EENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVV 122
EE SFD AI GC GVFH+ASP ++ S DP+ E+++PA+ G L+++++C K + R+V
Sbjct: 248 EEGSFDEAIKGCSGVFHVASP-MDFNSKDPENEVIKPAINGVLDIMKACLKAKTVRRLV 421
>TC17328 weakly similar to UP|Q9FGH3 (Q9FGH3) Dihydroflavonol
4-reductase-like (Cinnamoyl-CoA reductase-like protein),
partial (21%)
Length = 479
Score = 115 bits (289), Expect = 8e-27
Identities = 59/139 (42%), Positives = 87/139 (62%), Gaps = 1/139 (0%)
Frame = +3
Query: 51 ATERLKLVQADLMEENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLR 110
A R+ + AD+++ + AI C GVFH+ASP DP ++++PAVQGTLNVL
Sbjct: 66 AHSRITIFPADILDSAAVSRAIHNCSGVFHVASPCTLDDPEDPHKDLIQPAVQGTLNVLE 245
Query: 111 SCRKNPALVRVVLASSSSAVRVRADFDPNIPIDESSWSSLELCEKLQAWYPMSKTLAEKA 170
+ K + RVVL SS SA+ ++ +DESSW+ +E CE + WYP+SKT AE+A
Sbjct: 246 AA-KRAGVRRVVLTSSISALVPNPNWPEGRAVDESSWTDVEFCESRRKWYPVSKTAAERA 422
Query: 171 AWDYC-KENGIDLVTILPS 188
AW++ K+ G ++V I P+
Sbjct: 423 AWEFKEKQGGAEVVAIHPA 479
>TC16167 similar to UP|Q9FGH3 (Q9FGH3) Dihydroflavonol 4-reductase-like
(Cinnamoyl-CoA reductase-like protein), partial (44%)
Length = 535
Score = 115 bits (288), Expect = 1e-26
Identities = 62/148 (41%), Positives = 91/148 (60%)
Frame = +2
Query: 1 MEHKVCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQA 60
M VCVTG SG + SWL+ LL GY V TV++L + + +HL LEGA+ RL+L Q
Sbjct: 92 MSEHVCVTGGSGCIGSWLVHLLLDRGYTVHATVQNLNDETETKHLQSLEGASTRLRLFQI 271
Query: 61 DLMEENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVR 120
DL+ ++ A+ GC GVFH+ASP + +DP+ E+L+PA++GT NVL + K + R
Sbjct: 272 DLLHYDTVLAAVNGCAGVFHLASPCIVDQVHDPQKELLDPAIKGTENVL-AAAKEAGVRR 448
Query: 121 VVLASSSSAVRVRADFDPNIPIDESSWS 148
VV+ SS SA+ + ++ E W+
Sbjct: 449 VVVTSSVSAITPSPSWPSDVVKREDCWT 532
>TC8549 weakly similar to UP|Q8LEH3 (Q8LEH3) Cinnamoyl-CoA reductase-like
protein, partial (17%)
Length = 758
Score = 103 bits (258), Expect = 3e-23
Identities = 59/194 (30%), Positives = 106/194 (54%), Gaps = 1/194 (0%)
Frame = +3
Query: 3 HKVCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADL 62
H VCV ASG L L+++LL GY V +++ G +++ + ++ +++LK+ ++D
Sbjct: 150 HTVCVMDASGQLGFRLVQKLLQRGYTVHASIQKYGVDKEMFN--EISADSDKLKVFRSDP 323
Query: 63 MEENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVV 122
+ +S +A+ GC G+F+ P + D +E V+ NVL +C + + +VV
Sbjct: 324 FDYHSLIDALKGCSGLFYTFEPPFDQPDYDEFMADVE--VRAAHNVLEACAQTETMDKVV 497
Query: 123 LASSSSAVRVRADFDP-NIPIDESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCKENGID 181
S+++A R D + +DE WS + C K + W+ MSKTLAEK AW + G++
Sbjct: 498 FTSAATATVWREDRKTMELDLDERHWSDVNFCRKFKLWHGMSKTLAEKTAWALAMDTGVN 677
Query: 182 LVTILPSFIIGPNL 195
+V+I ++ +L
Sbjct: 678 MVSINSGLVMSHDL 719
>TC16256 similar to UP|Q7Y0H8 (Q7Y0H8) Cinnamoyl CoA reductase, partial
(32%)
Length = 475
Score = 95.9 bits (237), Expect = 8e-21
Identities = 57/105 (54%), Positives = 66/105 (62%)
Frame = +3
Query: 5 VCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLME 64
VCVTGA GF+ASWL+K LL GY V GTVR+ K HL +LEGA ERL L +ADL +
Sbjct: 144 VCVTGAGGFIASWLVKLLLERGYIVRGTVRN-PDDPKNGHLKELEGAKERLTLHKADLFD 320
Query: 65 ENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVL 109
+S GC GVFH ASPV D E+LEPAV G NV+
Sbjct: 321 LDSLKAVFHGCDGVFHTASPV-----TDNPEEMLEPAVNGARNVV 440
>AV774784
Length = 464
Score = 57.0 bits (136), Expect(2) = 7e-16
Identities = 30/72 (41%), Positives = 47/72 (64%)
Frame = +2
Query: 61 DLMEENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVR 120
+L EN FD I G VF +A+PV N S DP+ ++++PA++G LNVL+SC + + R
Sbjct: 233 ELTVENDFDTPIAGSDLVFQLATPV-NFASEDPENDMIKPAIKGVLNVLKSCAR-AKVKR 406
Query: 121 VVLASSSSAVRV 132
V+ SS+ +V +
Sbjct: 407 VIFTSSAVSVTI 442
Score = 42.7 bits (99), Expect(2) = 7e-16
Identities = 22/46 (47%), Positives = 29/46 (62%)
Frame = +1
Query: 4 KVCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLE 49
+ CV G +GF+AS LIK+LL GY V TVRD +K+ L L+
Sbjct: 64 EACVIGGTGFVASLLIKQLLEKGYAVNTTVRDPDNHKKISQLLALQ 201
>TC15771 similar to UP|AAR27015 (AAR27015) Dihydroflavonal-4-reductase 2,
partial (41%)
Length = 907
Score = 75.9 bits (185), Expect = 9e-15
Identities = 43/140 (30%), Positives = 72/140 (50%), Gaps = 3/140 (2%)
Frame = +3
Query: 190 IIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDVALCHILLYENKASDGRYL 249
++GP L + + L L G + + YVH+DD+ L HI L+EN + GRY+
Sbjct: 3 VVGPFLMPTMPPSLITALSLITGNEAHYSIIKQGQYVHLDDLCLAHIFLFENPKAQGRYM 182
Query: 250 CSSKIMDNDDLVGMLANRYPGFPIPKRFKKL--DRPHYELNTGKLESLGFKFK-SVEEMF 306
CS+ ++ M+ +YP F +P +FK + + + ++ K+ LGFKFK S+E+M+
Sbjct: 183 CSAYEATIHEVARMINKKYPEFNVPTKFKDIPDELDIIKFSSKKITDLGFKFKYSLEDMY 362
Query: 307 DDCFASFVEQGHLTTLPHQP 326
+ E+G L P
Sbjct: 363 TGAVETCREKGLLPKTAETP 422
>TC8762 similar to UP|O65880 (O65880) Cinnamoyl CoA reductase , partial
(32%)
Length = 544
Score = 73.2 bits (178), Expect = 6e-14
Identities = 39/104 (37%), Positives = 56/104 (53%), Gaps = 3/104 (2%)
Frame = +1
Query: 225 YVHIDDVALCHILLYENKASDGRYLCSSKIMDNDDLVGMLANRYPGFPIPKRFKKLDRPH 284
Y + DVAL HIL+YE + GRY+CS + +LV +LA +P +P+P + K P
Sbjct: 19 YADVRDVALAHILVYERPEASGRYICSESSLHRGELVEILAKHFPEYPMPTKCKDEKNPR 198
Query: 285 ---YELNTGKLESLGFKFKSVEEMFDDCFASFVEQGHLTTLPHQ 325
Y + KL+ LG +F V + S E+GHL T+P Q
Sbjct: 199 AKPYIFSNPKLKDLGLEFTPVSHSLYETVKSLQEKGHL-TIPKQ 327
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,382,196
Number of Sequences: 28460
Number of extensions: 90434
Number of successful extensions: 490
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 452
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 454
length of query: 330
length of database: 4,897,600
effective HSP length: 91
effective length of query: 239
effective length of database: 2,307,740
effective search space: 551549860
effective search space used: 551549860
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC138130.14


