

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138017.6 - phase: 0
(300 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
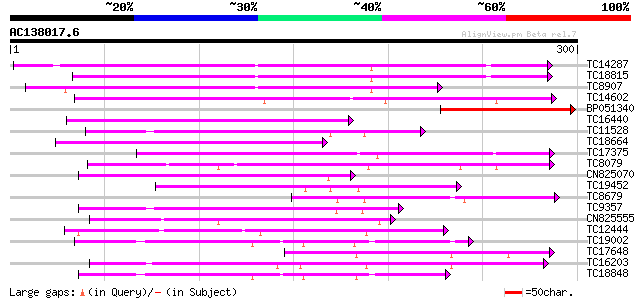
Score E
Sequences producing significant alignments: (bits) Value
TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein k... 182 5e-47
TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein ki... 161 1e-40
TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein ki... 151 1e-37
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 151 1e-37
BP051340 120 4e-28
TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinas... 112 1e-25
TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial... 109 5e-25
TC18664 similar to UP|Q944I6 (Q944I6) At2g30360/T9D9.17, partial... 104 2e-23
TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corn... 102 6e-23
TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete 101 1e-22
CN825070 100 3e-22
TC19452 homologue to UP|Q8S9J9 (Q8S9J9) At1g14000/F7A19_9, parti... 93 5e-20
TC8679 homologue to UP|Q43466 (Q43466) Protein kinase 3 , partia... 93 5e-20
TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein k... 93 6e-20
CN825555 91 3e-19
TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase iso... 90 5e-19
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 88 2e-18
TC17648 similar to UP|Q9AYN9 (Q9AYN9) NQK1 MAPKK, partial (51%) 88 2e-18
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 86 8e-18
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 84 3e-17
>TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein kinase-like
protein (CBL-interacting protein kinase 5), partial
(75%)
Length = 1999
Score = 182 bits (463), Expect = 5e-47
Identities = 103/291 (35%), Positives = 165/291 (56%), Gaps = 6/291 (2%)
Frame = +3
Query: 3 IATETQPQPHQQHHTASSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHI 62
+AT P P ++S+ V ++ R I N +++G+ LG+G F VY R +N
Sbjct: 339 LATPPPPPPPPPPPSSSTMVP---ENPRNIIFNKYEMGRVLGQGNFAKVYHGRNLATNEN 509
Query: 63 VALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGEL 122
VA+KV+ K +L++ ++ Q++REV + +RHPHI+ L + +++L++EY GEL
Sbjct: 510 VAIKVIKKEKLKKDRLVKQIKREVSVMRLVRHPHIVELKEVMATKGKIFLVMEYVKGGEL 689
Query: 123 YKELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWS 182
+ ++ K K +E A Y L A+ +CH + V HRD+KPENLL+ +LK++DFG S
Sbjct: 690 FTKVNKGK-LNEDDARKYFQQLISAVDFCHSRGVTHRDLKPENLLLDENEDLKVSDFGLS 866
Query: 183 VHTFNRRR-----TMCGTLDYLPPEMVESVEHDAS-VDIWSLGVLCYEFLYGVPPFEAKE 236
RR T CGT Y+ PE+++ +D S DIWS GV+ Y L G PF+ +
Sbjct: 867 ALPEQRRDDGMLVTPCGTPAYVAPEVLKKKGYDGSKADIWSCGVILYALLSGYLPFQGEN 1046
Query: 237 HSDTYRRIIQVDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPW 287
YR+ + + +FP +S AK+LIS +LV D +R + +++ PW
Sbjct: 1047VMRIYRKAFKAEYEFP--EWISPQAKNLISNLLVADPEKRYSIPEIISDPW 1193
>TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein kinase
CIPK25, partial (58%)
Length = 979
Score = 161 bits (408), Expect = 1e-40
Identities = 88/260 (33%), Positives = 146/260 (55%), Gaps = 6/260 (2%)
Frame = +2
Query: 34 LNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLR 93
+ +++G+ LG+G VY A+E TS VA+KV+ K+++++ + Q++RE+ I +R
Sbjct: 29 IGKYEMGRVLGKGTLAKVYFAKEITSGEGVAIKVMSKARIKKEGMMDQIKREISIMRLVR 208
Query: 94 HPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHG 153
HP+I+ L + +++ I+EY GEL+ ++ K K + A Y L A+ YCH
Sbjct: 209 HPNIVNLKEVMATKTKIFFIMEYIRGGELFAKVAKGK-LKDDLARRYFQQLISAVDYCHS 385
Query: 154 KHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRRR-----TMCGTLDYLPPEMVESVE 208
+ V HRD+KPENLL+ LK++DFG S R+ T CGT Y+ PE++
Sbjct: 386 RGVSHRDLKPENLLLDENENLKVSDFGLSGLPEQLRQDGLLHTQCGTPAYVAPEVLRKKG 565
Query: 209 HDA-SVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLISQ 267
+D D WS GV+ Y L G PF+ + Y ++++ + +FP P S +K LIS+
Sbjct: 566 YDGFKTDTWSCGVILYALLAGCLPFQHENLMTMYNKVLRAEFQFP--PWFSPESKKLISK 739
Query: 268 MLVKDSSERLPLHKLLEHPW 287
+LV D + R+ + ++ W
Sbjct: 740 ILVADPNRRITISSIMRVSW 799
>TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein kinase-like
protein, partial (44%)
Length = 941
Score = 151 bits (382), Expect = 1e-37
Identities = 84/231 (36%), Positives = 133/231 (57%), Gaps = 10/231 (4%)
Frame = +3
Query: 9 PQPHQQHHTASSEVSGSAKD----QRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVA 64
PQP ++ T + + ++ +R + N ++IGK LG+G F VY R +N VA
Sbjct: 246 PQPTTRNKTINMDQRQQSRTMRAAKRNILFNKYEIGKILGQGNFAKVYHGRNMETNESVA 425
Query: 65 LKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYK 124
+KV+ K +L++ ++ Q++REV + +RHPHI+ L + ++++++EY GEL+
Sbjct: 426 IKVIKKERLKKERLVKQIKREVSVMRLVRHPHIVELKEVMATKTKIFMVVEYVKGGELFA 605
Query: 125 ELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVH 184
+L K K +E A Y L A+ +CH + V HRD+KPENLL+ +LK++DFG S
Sbjct: 606 KLTKGK-MTEVAARKYFQQLISAVDFCHSRGVTHRDLKPENLLLDDNEDLKVSDFGLSSL 782
Query: 185 TFNRRR-----TMCGTLDYLPPEMVESVEHDAS-VDIWSLGVLCYEFLYGV 229
RR T CGT Y+ PE+++ +D S DIWS GV+ Y L G+
Sbjct: 783 PEQRRSDGMLLTPCGTPAYVAPEVLKKKGYDGSKADIWSCGVILYALLCGI 935
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 151 bits (381), Expect = 1e-37
Identities = 89/263 (33%), Positives = 140/263 (52%), Gaps = 8/263 (3%)
Frame = +2
Query: 35 NDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLR- 93
+ + + + +GRG+FG ++ S A+K++ KS L S H L E + S L
Sbjct: 62 SQYQLCEEIGRGRFGTIFRCFHPISTDTFAVKLIDKSLLADSTDRHCLENEPKYMSLLSP 241
Query: 94 HPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFS--ERRAATYVASLARALIYC 151
HP+IL+++ F D + +++E L + S E AA + L A+ +C
Sbjct: 242 HPNILQIFDVFEDDDVLSMVIELCQPLTLLDRIVAANGTSIPEVEAAGLMKQLLEAVAHC 421
Query: 152 HGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRRRTMCGTLD---YLPPEMVESVE 208
H V HRD+KP+N+L G G+LK+ADFG S F R M G + Y+ PE++ E
Sbjct: 422 HRLGVAHRDVKPDNVLFGGGGDLKLADFG-SAEWFGDGRRMSGVVGTPYYVAPEVLMGRE 598
Query: 209 HDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPI--VSSAAKDLIS 266
+ VD+WS GV+ Y L G PPF ++ + +I+ +L+FP + VS AAKDL+
Sbjct: 599 YGEKVDVWSCGVILYIMLSGTPPFYGDSAAEIFEAVIRGNLRFPSRIFRNVSPAAKDLLR 778
Query: 267 QMLVKDSSERLPLHKLLEHPWIV 289
+M+ +D S R+ + L HPW +
Sbjct: 779 KMICRDPSNRISAEQALRHPWFL 847
>BP051340
Length = 537
Score = 120 bits (300), Expect = 4e-28
Identities = 58/71 (81%), Positives = 61/71 (85%)
Frame = -3
Query: 229 VPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPWI 288
V P D YRRI+QVDLKFPPKPIVSSAAKDLISQMLVKDS +RLPLHKLLEHPWI
Sbjct: 502 VNPTFLXRSEDAYRRIVQVDLKFPPKPIVSSAAKDLISQMLVKDSCQRLPLHKLLEHPWI 323
Query: 289 VQNAEPSGIYR 299
VQNAEPSG+YR
Sbjct: 322 VQNAEPSGVYR 290
>TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinase, partial
(30%)
Length = 549
Score = 112 bits (279), Expect = 1e-25
Identities = 58/153 (37%), Positives = 92/153 (59%), Gaps = 1/153 (0%)
Frame = +1
Query: 31 RWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQS 90
R IL + + + LGRG F VY A T VA+K++ KS+ + +E ++ RE++
Sbjct: 73 RIILGKYQLTRFLGRGNFAKVYQAVSLTDGTTVAVKMIDKSKTVDASMEPRIVREIDAMR 252
Query: 91 HLRH-PHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALI 149
L+H P+IL+++ + ++YLI++YA GEL+ ++ + E A Y L AL
Sbjct: 253 RLQHHPNILKIHEVMATRTKIYLIVDYAGGGELFSKISRRGRLPEPLARRYFQQLVSALC 432
Query: 150 YCHGKHVIHRDIKPENLLIGAQGELKIADFGWS 182
+CH V HRD+KP+NLL+ A+G LK++DFG S
Sbjct: 433 FCHRNGVAHRDLKPQNLLLDAEGNLKVSDFGLS 531
>TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial (56%)
Length = 640
Score = 109 bits (273), Expect = 5e-25
Identities = 63/185 (34%), Positives = 99/185 (53%), Gaps = 5/185 (2%)
Frame = +3
Query: 41 KPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRL 100
K LG G FG LAR+K + +VA+K + + + +++ ++RE+ LRHP+I+R
Sbjct: 90 KELGSGNFGVARLARDKNTGELVAVKYIERGK----KIDENVQREIINHRSLRHPNIIRF 257
Query: 101 YGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHGKHVIHRD 160
+ ++LEYA GEL++ + FSE A + L + YCH + HRD
Sbjct: 258 KEVLLTPTHLAIVLEYASGGELFERICSAGRFSEDEARYFFQQLISGVSYCHSMEICHRD 437
Query: 161 IKPENLLI--GAQGELKIADFGWSVHTF--NRRRTMCGTLDYLPPEMVESVEHDASV-DI 215
+K EN L+ LKI DFG+S ++ ++ GT Y+ PE++ E+D V D+
Sbjct: 438 LKLENTLLDGNPSPRLKICDFGYSKSAILHSQPKSTVGTPAYIAPEVLSRKEYDGKVADV 617
Query: 216 WSLGV 220
WS GV
Sbjct: 618 WSCGV 632
>TC18664 similar to UP|Q944I6 (Q944I6) At2g30360/T9D9.17, partial (34%)
Length = 643
Score = 104 bits (260), Expect = 2e-23
Identities = 53/144 (36%), Positives = 84/144 (57%)
Frame = +3
Query: 25 SAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRR 84
+A + + +++G+ LG G F VY AR + VA+KV+ K ++ + + ++R
Sbjct: 57 AAAESTTVLFGKYEVGRLLGCGAFAKVYHARNIETGQSVAVKVINKKKVIGTGLTGHVKR 236
Query: 85 EVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASL 144
EV I S LRHP+I+RL+ + ++Y ++E+A GEL+ + FSE A Y L
Sbjct: 237 EVSIMSRLRHPNIVRLHEVLATKTKIYFVMEFAKGGELFARISTKGRFSEDLARRYFQQL 416
Query: 145 ARALIYCHGKHVIHRDIKPENLLI 168
A+ YCH + V HRD+KPENLL+
Sbjct: 417 ISAVGYCHSRGVFHRDLKPENLLL 488
>TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corniculatus
var. japonicus]
Length = 1422
Score = 102 bits (255), Expect = 6e-23
Identities = 70/226 (30%), Positives = 109/226 (47%), Gaps = 5/226 (2%)
Frame = +3
Query: 68 LFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQ 127
+F + QL +E+ + + HP+I++ YG ++ + + LEY G ++K LQ
Sbjct: 30 VFSDDKTSKECLKQLNQEINLLNQFSHPNIVQYYGSELGEESLSVYLEYVSGGSIHKLLQ 209
Query: 128 KCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFN 187
+ F E Y + L Y H ++ +HRDIK N+L+ GE+K+ADFG S H N
Sbjct: 210 EYGAFKEPVIQNYTRQIVSGLAYLHSRNTVHRDIKGANILVDPNGEIKLADFGMSKH-IN 386
Query: 188 RRRTMC---GTLDYLPPEMVESVE-HDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRR 243
+M G+ ++ PE+V + + VDI SLG E PP+ E +
Sbjct: 387 SAASMLSFKGSPYWMAPEVVMNTNGYGLPVDISSLGCTILEMATSKPPWSQFEGVAAIFK 566
Query: 244 I-IQVDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPWI 288
I D+ P+ + S AK+ I Q L +D R LL HP+I
Sbjct: 567 IGNSKDMPEIPEHL-SDDAKNFIKQCLQRDPLARPTAQSLLNHPFI 701
>TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete
Length = 1550
Score = 101 bits (252), Expect = 1e-22
Identities = 75/284 (26%), Positives = 133/284 (46%), Gaps = 37/284 (13%)
Frame = +3
Query: 42 PLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLY 101
P+GRG +G V +N +VA+K + + + LR E+++ HL H +++ L
Sbjct: 306 PIGRGAYGIVCSLLNTETNELVAVKKIANAFDNHMDAKRTLR-EIKLLRHLDHENVIALR 482
Query: 102 GYFYDQKR-----VYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHGKHV 156
R VY+ E +L++ ++ + SE ++ + R L Y H ++
Sbjct: 483 DVIPPPLRREFTDVYITTELMDT-DLHQIIRSNQGLSEEHCQYFLYQVLRGLKYIHSANI 659
Query: 157 IHRDIKPENLLIGAQGELKIADFGWSVHTFNR--RRTMCGTLDYLPPE-MVESVEHDASV 213
IHRD+KP NLL+ + +LKI DFG + T T Y PE ++ S ++ +++
Sbjct: 660 IHRDLKPSNLLLNSNCDLKIIDFGLARPTVENDFMTEYVVTRWYRAPELLLNSSDYTSAI 839
Query: 214 DIWSLGVLCYEFLYGVPPFEAKEH-----------------------SDTYRRIIQVDLK 250
D+WS+G + E + P F K+H +D RR I+ +
Sbjct: 840 DVWSVGCIFMELMNKKPLFPGKDHVHQLRLLTELLGTPTEADLGLVKNDDVRRYIRQLPQ 1019
Query: 251 FPPKPI------VSSAAKDLISQMLVKDSSERLPLHKLLEHPWI 288
+P +P+ V A DL+ +ML D ++R+ + + L HP++
Sbjct: 1020YPRQPLTKVFPHVHPMAMDLVDKMLTIDPTKRITVEQALAHPYL 1151
>CN825070
Length = 634
Score = 100 bits (249), Expect = 3e-22
Identities = 52/151 (34%), Positives = 85/151 (55%), Gaps = 4/151 (2%)
Frame = +3
Query: 37 FDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHL-RHP 95
+ G+ LG+G+FG YL R ++ A K + K +L + + RE++I HL HP
Sbjct: 129 YTFGRKLGQGQFGTTYLCRHNSTGCTFACKSIPKRKLLCKEDYDDVWREIQIMHHLSEHP 308
Query: 96 HILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHGKH 155
H++R++G + D V++++E GEL+ + + +SER AA + ++ + CH
Sbjct: 309 HVVRIHGTYEDAASVHIVMEICEGGELFDRIVQKGQYSEREAAKLIRTIVEVVEACHSLG 488
Query: 156 VIHRDIKPENLL---IGAQGELKIADFGWSV 183
V+HRD+KPEN L + +LK DFG SV
Sbjct: 489 VMHRDLKPENFLFDTVEEDAKLKTTDFGLSV 581
>TC19452 homologue to UP|Q8S9J9 (Q8S9J9) At1g14000/F7A19_9, partial (41%)
Length = 562
Score = 93.2 bits (230), Expect = 5e-20
Identities = 55/173 (31%), Positives = 89/173 (50%), Gaps = 11/173 (6%)
Frame = +3
Query: 78 VEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRA 137
V R EV + LRHP+I++ G ++K + LI EY G+L++ L++ S A
Sbjct: 27 VIQDFRHEVNLLVKLRHPNIVQFLGAVTERKPLMLITEYLRGGDLHQYLKEKGSLSPSTA 206
Query: 138 ATYVASLARALIYCHGKH--VIHRDIKPENLLI--GAQGELKIADFGWSV-------HTF 186
+ + R + Y H + +IHRD+KP N+L+ + LK+ DFG S H
Sbjct: 207 INFSMDIVRGMAYLHNEPNVIIHRDLKPRNVLLVNSSADHLKVGDFGLSKLITVQNSHDV 386
Query: 187 NRRRTMCGTLDYLPPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSD 239
+ G+ Y+ PE+ + ++D VD++S ++ YE L G PPF + E D
Sbjct: 387 YKMTGETGSYRYMAPEVFKHRKYDKKVDVYSFAMILYEMLEGEPPFASYEPYD 545
>TC8679 homologue to UP|Q43466 (Q43466) Protein kinase 3 , partial (70%)
Length = 972
Score = 93.2 bits (230), Expect = 5e-20
Identities = 54/153 (35%), Positives = 82/153 (53%), Gaps = 11/153 (7%)
Frame = +2
Query: 150 YCHGKHVIHRDIKPENLLIGAQG--ELKIADFGWSVHTF--NRRRTMCGTLDYLPPEMVE 205
YCH + HRD+K EN L+ LKI DFG+S + +R ++ GT Y+ PE++
Sbjct: 26 YCHALQICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLS 205
Query: 206 SVEHDASV-DIWSLGVLCYEFLYGVPPFEAKEHSD------TYRRIIQVDLKFPPKPIVS 258
E+D + D+WS GV Y L G PFE +H D T +RI+ V K P +S
Sbjct: 206 RREYDGKLADVWSCGVTLYVMLVGAYPFE--DHDDPRNFRKTIQRIMAVQYKIPDYVHIS 379
Query: 259 SAAKDLISQMLVKDSSERLPLHKLLEHPWIVQN 291
+ L+S++ V + R+ L ++ HPW ++N
Sbjct: 380 QDCRHLLSRIFVANPLRRISLKEIKNHPWFLKN 478
>TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein kinase,
41K) , partial (71%)
Length = 1181
Score = 92.8 bits (229), Expect = 6e-20
Identities = 52/176 (29%), Positives = 92/176 (51%), Gaps = 4/176 (2%)
Frame = +1
Query: 37 FDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPH 96
+D+ + +G G FG L ++K + +VA+K + + +++ ++RE+ LRHP+
Sbjct: 361 YDLVRDIGSGNFGVARLMQDKQTKELVAVKYIERGD----KIDENVKREIINHRSLRHPN 528
Query: 97 ILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHGKHV 156
I+R + +++EYA GEL++ + F+E A + L + YCH V
Sbjct: 529 IVRFKEVILTPTHLAIVMEYASGGELFERICNAGRFTEDEARFFFQQLISGVSYCHAMQV 708
Query: 157 IHRDIKPENLLIGAQ--GELKIADFGWSVHT--FNRRRTMCGTLDYLPPEMVESVE 208
HRD+K EN L+ LKI DFG+S + ++ ++ GT Y+ PE++ E
Sbjct: 709 CHRDLKLENTLLDGSPTPRLKICDFGYSKSSVLHSQPKSTVGTPAYIAPEVLSKQE 876
>CN825555
Length = 714
Score = 90.5 bits (223), Expect = 3e-19
Identities = 60/168 (35%), Positives = 88/168 (51%), Gaps = 6/168 (3%)
Frame = +1
Query: 43 LGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYG 102
+G+G + +VY AR+ + IVALK + L+ V+ + RE+ I L HP++L+L G
Sbjct: 202 IGQGTYSNVYKARDTLTGKIVALKKVRFDNLEPESVKF-MAREILILRRLDHPNVLKLEG 378
Query: 103 YFYDQKR--VYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHGKHVIHRD 160
+ +YL+ EY F+E + Y+ L L +CH +HV+HRD
Sbjct: 379 LVTSRMSCSLYLVFEYMVHDLAGLATNPAIKFTESQVKCYMHQLFTGLEHCHNRHVLHRD 558
Query: 161 IKPENLLIGAQGELKIADFGW-SVHTFNRRRTMCG---TLDYLPPEMV 204
IK NLLI +G LKIADFG S + + M TL Y PPE++
Sbjct: 559 IKGSNLLIDNEGVLKIADFGLASFFDPDHKHPMTSRVVTLWYRPPELL 702
>TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase isolog,
partial (37%)
Length = 709
Score = 89.7 bits (221), Expect = 5e-19
Identities = 60/215 (27%), Positives = 108/215 (49%), Gaps = 12/215 (5%)
Frame = +2
Query: 30 RRWILN--DFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVE 87
R + LN D+++ + +G G VY A + VA+K + + + + +RRE +
Sbjct: 62 RTYSLNSGDYNLLEEVGYGASATVYRAVFLPRDEEVAVKCVDLDRCNANLED--IRREAQ 235
Query: 88 IQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKY---FSERRAATYVASL 144
S + H +++R + F +R+++++ + +G L K Y F E + +
Sbjct: 236 TMSLIDHRNVVRAHWSFVVDRRLWVVMPFMAQGSCL-HLMKVAYPDGFEEEAIGSILKET 412
Query: 145 ARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFN------RRRTMCGTLDY 198
+AL Y H IHRD+K N+L+G+ G++K+ADFG S F+ R T GT +
Sbjct: 413 LKALEYLHRHGHIHRDVKAGNILLGSDGQVKLADFGVSASMFDAGERQRNRNTFVGTPCW 592
Query: 199 LPPEMVE-SVEHDASVDIWSLGVLCYEFLYGVPPF 232
+ PE+++ ++ D+WS G+ E +G PF
Sbjct: 593 MAPEVLQPGTGYNFKADVWSFGITALELAHGHAPF 697
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 88.2 bits (217), Expect = 2e-18
Identities = 65/226 (28%), Positives = 108/226 (47%), Gaps = 15/226 (6%)
Frame = +1
Query: 35 NDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRH 94
N+F+ LG G FG VY + + I ++ ++ ++ + + EVEI + +RH
Sbjct: 58 NNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRL----KVWSNKADMEFAVEVEILARVRH 225
Query: 95 PHILRLYGYFYDQKRVYLILEYAPKGELYKELQ-----KCKYFSERRAATYVASLARALI 149
++L L GY + + ++ +Y P L L +C RR + S A ++
Sbjct: 226 KNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIGS-AEGIV 402
Query: 150 YCHGK---HVIHRDIKPENLLIGAQGELKIADFGW-------SVHTFNRRRTMCGTLDYL 199
Y H + H+IHRDIK N+L+ + + ++ADFG+ + H R + GTL YL
Sbjct: 403 YLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVTTRVK---GTLGYL 573
Query: 200 PPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRII 245
PE + + D++S G+L E G P E + S T +R I
Sbjct: 574 APEYAMLGKANECCDVFSFGILLLELASGKKPLE--KLSSTVKRSI 705
>TC17648 similar to UP|Q9AYN9 (Q9AYN9) NQK1 MAPKK, partial (51%)
Length = 741
Score = 88.2 bits (217), Expect = 2e-18
Identities = 53/154 (34%), Positives = 82/154 (52%), Gaps = 11/154 (7%)
Frame = +3
Query: 146 RALIYCHG-KHVIHRDIKPENLLIGAQGELKIADFGWS---VHTFNRRRTMCGTLDYLPP 201
+ L+Y H +HVIHRD KP +LL+ QGE+KI DFG S + +R T GT +Y+ P
Sbjct: 6 QGLVYLHNERHVIHRDSKPSHLLVNHQGEVKITDFGVSAMLATSMGQRDTFVGTYNYMSP 185
Query: 202 EMVESVEHDASVDIWSLGVLCYEFLYGVPPF---EAKEHSDTYRRIIQVDLKFPPKPIVS 258
+ +D S DIWSLG++ E G P+ E ++ ++ ++ ++ PP S
Sbjct: 186 ARISGSTYDYSSDIWSLGMVVLECAIGRFPYIQSEDQQAWPSFYELLAAIVESPPPSAPS 365
Query: 259 SAAK----DLISQMLVKDSSERLPLHKLLEHPWI 288
+S + KD +RL +LL+HP+I
Sbjct: 366 DQFSPEFCSFVSSCIQKDPQDRLTSLELLDHPFI 467
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 85.9 bits (211), Expect = 8e-18
Identities = 69/271 (25%), Positives = 114/271 (41%), Gaps = 28/271 (10%)
Frame = +2
Query: 43 LGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYG 102
+G+G G VY + +++ + + ++ R E+E +RH +I+RL G
Sbjct: 2210 IGKGGAGIVYRGSMPNGTDVAIKRLVGQGS---GRNDYGFRAEIETLGKIRHRNIMRLLG 2380
Query: 103 YFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATY--VASLARALIYCH---GKHVI 157
Y ++ L+ EY P G L + L K R Y AR L Y H +I
Sbjct: 2381 YVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLII 2560
Query: 158 HRDIKPENLLIGAQGELKIADFGWSVHTFN-----RRRTMCGTLDYLPPEMVESVEHDAS 212
HRD+K N+L+ A E +ADFG + ++ ++ G+ Y+ PE +++ D
Sbjct: 2561 HRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEK 2740
Query: 213 VDIWSLGVLCYEFLYGVPPF------------------EAKEHSDTYRRIIQVDLKFPPK 254
D++S GV+ E + G P E + SDT + VD +
Sbjct: 2741 SDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGY 2920
Query: 255 PIVSSAAKDLISQMLVKDSSERLPLHKLLEH 285
P+ S I+ M VK+ P + + H
Sbjct: 2921 PLTSVIHMFNIAMMCVKEMGPARPTMREVVH 3013
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 84.0 bits (206), Expect = 3e-17
Identities = 63/212 (29%), Positives = 96/212 (44%), Gaps = 15/212 (7%)
Frame = +2
Query: 37 FDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPH 96
F LG G FG VY R I K+ + S+ E + EVE+ +RH +
Sbjct: 302 FSDDNKLGEGGFGSVYWGRTSDGLQIAVKKL----KAMNSKAEMEFAVEVEVLGRVRHKN 469
Query: 97 ILRLYGYFYDQKRVYLILEYAPKGELYKELQ-----KCKYFSERRAATYVASLARALIYC 151
+L L GY + ++ +Y P L L + + ++R + S A ++Y
Sbjct: 470 LLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIAIGS-AEGILYL 646
Query: 152 HGK---HVIHRDIKPENLLIGAQGELKIADFGWS-------VHTFNRRRTMCGTLDYLPP 201
H + H+IHRDIK N+L+ + E +ADFG++ H R + GTL YL P
Sbjct: 647 HHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSHMTTRVK---GTLGYLAP 817
Query: 202 EMVESVEHDASVDIWSLGVLCYEFLYGVPPFE 233
E + S D++S G+L E + G P E
Sbjct: 818 EYAMWGKVSESCDVYSFGILLLELVTGRKPIE 913
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.137 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,883,520
Number of Sequences: 28460
Number of extensions: 86666
Number of successful extensions: 702
Number of sequences better than 10.0: 215
Number of HSP's better than 10.0 without gapping: 632
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 638
length of query: 300
length of database: 4,897,600
effective HSP length: 90
effective length of query: 210
effective length of database: 2,336,200
effective search space: 490602000
effective search space used: 490602000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC138017.6


