

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138015.8 + phase: 0
(261 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
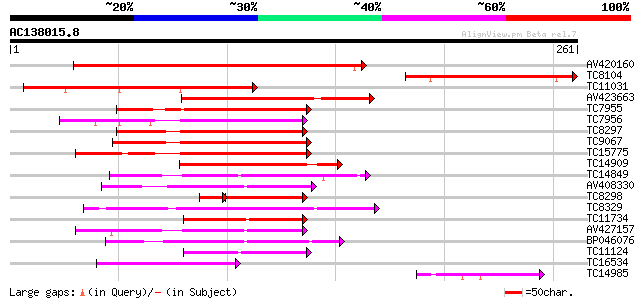
Score E
Sequences producing significant alignments: (bits) Value
AV420160 229 4e-61
TC8104 homologue to UP|WR65_ARATH (Q9LP56) Probable WRKY transcr... 119 4e-28
TC11031 homologue to UP|WR65_ARATH (Q9LP56) Probable WRKY transc... 114 2e-26
AV423663 109 5e-25
TC7955 similar to UP|WR11_ARATH (Q9SV15) Probable WRKY transcrip... 107 2e-24
TC7956 similar to GB|AAM61419.1|21537078|AY084855 putaive DNA-bi... 107 2e-24
TC8297 similar to UP|Q40828 (Q40828) WRKY3, partial (27%) 104 1e-23
TC9067 similar to UP|WR15_ARATH (O22176) Probable WRKY transcrip... 104 2e-23
TC15775 similar to UP|WR17_ARATH (Q9SJA8) Probable WRKY transcri... 103 3e-23
TC14909 similar to UP|WR21_ARATH (O04336) Probable WRKY transcri... 101 1e-22
TC14849 similar to UP|Q40090 (Q40090) SPF1 protein, partial (37%) 78 2e-15
AV408330 77 2e-15
TC8298 homologue to UP|Q9SXP4 (Q9SXP4) DNA-binding protein NtWRK... 62 1e-14
TC8329 similar to UP|BAD06717 (BAD06717) WRKY transcription fact... 75 1e-14
TC11734 similar to UP|WR49_ARATH (Q9FHR7) Probable WRKY transcri... 74 3e-14
AV427157 70 3e-13
BP046076 67 4e-12
TC11124 similar to UP|Q8H9D7 (Q8H9D7) WRKY-type DNA binding prot... 57 3e-09
TC16534 similar to UP|WR41_ARATH (Q8H0Y8) Probable WRKY transcri... 53 5e-08
TC14985 similar to UP|WR65_ARATH (Q9LP56) Probable WRKY transcri... 50 3e-07
>AV420160
Length = 417
Score = 229 bits (584), Expect = 4e-61
Identities = 108/136 (79%), Positives = 119/136 (87%), Gaps = 1/136 (0%)
Frame = +1
Query: 30 EDGPASPSSCEDTKIEESSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYG 89
E GPASPSS ED K E SPKKRREMKKRVVT+PI DV+GSKS+GETYPPSDSWAWRKYG
Sbjct: 4 EPGPASPSSGEDIKTEAPSPKKRREMKKRVVTLPIGDVDGSKSKGETYPPSDSWAWRKYG 183
Query: 90 QKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSHHSST 149
QKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPT L+VTY++EHNHSLP+PKSH S+
Sbjct: 184 QKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTKLMVTYSYEHNHSLPVPKSHSSAA 363
Query: 150 NAVTATVT-AAVDTPS 164
+A T + A DTP+
Sbjct: 364 SATATTSSDGAADTPA 411
>TC8104 homologue to UP|WR65_ARATH (Q9LP56) Probable WRKY transcription
factor 65 (WRKY DNA-binding protein 65), partial (12%)
Length = 604
Score = 119 bits (299), Expect = 4e-28
Identities = 62/90 (68%), Positives = 65/90 (71%), Gaps = 11/90 (12%)
Frame = +1
Query: 183 THPDFDLTGD---------HHAVFGWFDDIVSTGVLVSPICGGVEDVTLTMREEDESLFA 233
THPD + TGD HHA FGWFDD+ STGVL SPIC VEDV LTMREEDESLFA
Sbjct: 1 THPDLEFTGDSAVLLGHHHHHADFGWFDDVASTGVLESPICDEVEDVMLTMREEDESLFA 180
Query: 234 DLGELPECSTVFRQRNI--PSVFQYSGITG 261
DLGELPECS VFR+RNI S Q GITG
Sbjct: 181 DLGELPECSAVFRRRNILSTSTIQCDGITG 270
>TC11031 homologue to UP|WR65_ARATH (Q9LP56) Probable WRKY transcription
factor 65 (WRKY DNA-binding protein 65), partial (15%)
Length = 453
Score = 114 bits (284), Expect = 2e-26
Identities = 61/116 (52%), Positives = 76/116 (64%), Gaps = 8/116 (6%)
Frame = +3
Query: 7 NTKPLYVADREEPEPELK---LEPILEDGPASPSSCEDTKIEESSP----KKRREMKKRV 59
N + Y+ + E + E+ E I + P+S + D + SS + RR ++KRV
Sbjct: 105 NNRHRYINELLEEDAEVAHHHQENIADSPPSSTAFNIDGLVTSSSTSSAKRSRRAIQKRV 284
Query: 60 VTIPIADVEGSKSRGETY-PPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPAR 114
V IPI + EGS+ + E PPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPAR
Sbjct: 285 VQIPIKETEGSRLKAENNTPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPAR 452
>AV423663
Length = 447
Score = 109 bits (272), Expect = 5e-25
Identities = 52/89 (58%), Positives = 61/89 (68%)
Frame = +1
Query: 80 SDSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSL 139
SD WAWRKYGQKPIKGSPYPRGYYRCSSSKGC ARKQVER++ +P +VTY EHNH
Sbjct: 79 SDIWAWRKYGQKPIKGSPYPRGYYRCSSSKGCLARKQVERNKSEPAMFIVTYTGEHNHPA 258
Query: 140 PLPKSHHSSTNAVTATVTAAVDTPSPVES 168
P +H +S T +T + +S
Sbjct: 259 P---THRNSLAGSTRQKPLTPETATAGDS 336
>TC7955 similar to UP|WR11_ARATH (Q9SV15) Probable WRKY transcription
factor 11 (WRKY DNA-binding protein 11), partial (27%)
Length = 579
Score = 107 bits (268), Expect = 2e-24
Identities = 51/90 (56%), Positives = 63/90 (69%)
Frame = +3
Query: 50 KKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSK 109
KKR+ K+ V +P S+ PP D ++WRKYGQKPIKGSPYPRGYY+CS+ +
Sbjct: 18 KKRKNRVKKTVRVPAIS-----SKTADIPP-DEYSWRKYGQKPIKGSPYPRGYYKCSTVR 179
Query: 110 GCPARKQVERSRVDPTNLLVTYAHEHNHSL 139
GCPARK VER+ DPT L+VTY EH H+L
Sbjct: 180 GCPARKHVERATDDPTMLIVTYEGEHRHAL 269
>TC7956 similar to GB|AAM61419.1|21537078|AY084855 putaive DNA-binding
protein {Arabidopsis thaliana;} , partial (45%)
Length = 1257
Score = 107 bits (267), Expect = 2e-24
Identities = 58/129 (44%), Positives = 72/129 (54%), Gaps = 15/129 (11%)
Frame = +3
Query: 24 KLEPILEDGPASPSS------CEDTKIEESSP----KKRREMKKRVVTIP-----IADVE 68
K+ P L P S C D S P KKR+ K+ + +P IAD+
Sbjct: 471 KIGPFLTPAAKPPLSTSHRKRCHDAAALSSGPCHCSKKRKSRVKQTIRVPAVSSKIADI- 647
Query: 69 GSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLL 128
P D ++WRKYGQKPIKGSPYPRGYY+CS+ +GCPARK VER++ DP L+
Sbjct: 648 ----------PPDEYSWRKYGQKPIKGSPYPRGYYKCSTVRGCPARKHVERAQDDPNMLI 797
Query: 129 VTYAHEHNH 137
VTY EH H
Sbjct: 798 VTYEGEHRH 824
Score = 30.0 bits (66), Expect = 0.42
Identities = 20/50 (40%), Positives = 22/50 (44%), Gaps = 2/50 (4%)
Frame = +1
Query: 132 AHEHNHSLPLPKSHHS--STNAVTATVTAAVDTPSPVESPAATPQPEDRP 179
AH HNH+L L S N T T TPSP P TP P + P
Sbjct: 310 AHHHNHNLKLRSKA*L*ISLNPPTRT------TPSPSPPPTPTPPPSNPP 441
>TC8297 similar to UP|Q40828 (Q40828) WRKY3, partial (27%)
Length = 617
Score = 104 bits (260), Expect = 1e-23
Identities = 48/88 (54%), Positives = 60/88 (67%)
Frame = +1
Query: 50 KKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSK 109
KK++ KR + +P + + P+D + WRKYGQKPIKGSPYPRGYY+CSS K
Sbjct: 97 KKKKLRVKRTIRVPAISSKTADI------PTDDFQWRKYGQKPIKGSPYPRGYYKCSSEK 258
Query: 110 GCPARKQVERSRVDPTNLLVTYAHEHNH 137
GCPARK VER++ DP L+VTY EH H
Sbjct: 259 GCPARKHVERAQDDPNMLIVTYEGEHRH 342
>TC9067 similar to UP|WR15_ARATH (O22176) Probable WRKY transcription
factor 15 (WRKY DNA-binding protein 15), partial (31%)
Length = 660
Score = 104 bits (259), Expect = 2e-23
Identities = 50/92 (54%), Positives = 63/92 (68%)
Frame = +3
Query: 48 SPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSS 107
S K R+ KRVV +P ++ + P D ++WRKYGQKPIKGSP+PRGYY+CSS
Sbjct: 45 SQKSRKMRLKRVVRVPAISLKMADI------PPDDYSWRKYGQKPIKGSPHPRGYYKCSS 206
Query: 108 SKGCPARKQVERSRVDPTNLLVTYAHEHNHSL 139
+GCPARK VER+ D L+VTY EHNH+L
Sbjct: 207 VRGCPARKHVERALDDAAMLVVTYEGEHNHAL 302
>TC15775 similar to UP|WR17_ARATH (Q9SJA8) Probable WRKY transcription
factor 17 (WRKY DNA-binding protein 17), partial (53%)
Length = 1284
Score = 103 bits (257), Expect = 3e-23
Identities = 50/109 (45%), Positives = 69/109 (62%)
Frame = +3
Query: 31 DGPASPSSCEDTKIEESSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQ 90
D S + C TK ++ KK ++ V+ +AD+ P+D ++WRKYGQ
Sbjct: 630 DPSGSNTKCHCTKRRKNRVKKT--IRVPAVSSKVADI-----------PADEYSWRKYGQ 770
Query: 91 KPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSL 139
KPIKG+PYPRGYY+CS+ +GCPARK VER+ DP L+VTY EH+H +
Sbjct: 771 KPIKGTPYPRGYYKCSTVRGCPARKHVERATDDPAMLIVTYEDEHDHGI 917
>TC14909 similar to UP|WR21_ARATH (O04336) Probable WRKY transcription
factor 21 (WRKY DNA-binding protein 21), partial (22%)
Length = 590
Score = 101 bits (252), Expect = 1e-22
Identities = 48/75 (64%), Positives = 54/75 (72%)
Frame = +3
Query: 79 PSDSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHS 138
P D ++WRKYGQKPIKGSP+PRGYY+CSS +GCPARK VER +PT LLVTY EHNH
Sbjct: 57 PPDDYSWRKYGQKPIKGSPHPRGYYKCSSMRGCPARKHVERCLEEPTMLLVTYEGEHNH- 233
Query: 139 LPLPKSHHSSTNAVT 153
PK NA T
Sbjct: 234 ---PKIPTQPANA*T 269
>TC14849 similar to UP|Q40090 (Q40090) SPF1 protein, partial (37%)
Length = 1389
Score = 77.8 bits (190), Expect = 2e-15
Identities = 45/122 (36%), Positives = 64/122 (51%), Gaps = 2/122 (1%)
Frame = +2
Query: 47 SSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCS 106
S+P R + RVV +D++ D + WRKYGQK +KG+P PR YY+C
Sbjct: 332 SAPGSRTVREPRVVVQTTSDIDIL---------DDGYRWRKYGQKVVKGNPNPRSYYKC- 481
Query: 107 SSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPK--SHHSSTNAVTATVTAAVDTPS 164
+ GCP RK VER+ D ++ TY +HNH +P + +HS T + A+ PS
Sbjct: 482 THPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPLPNNTANAI-RPS 658
Query: 165 PV 166
V
Sbjct: 659 SV 664
>AV408330
Length = 316
Score = 77.4 bits (189), Expect = 2e-15
Identities = 38/99 (38%), Positives = 59/99 (59%)
Frame = +3
Query: 43 KIEESSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGY 102
K ++++ K++RE + +T + E D + WRKYGQK +K SP+PR Y
Sbjct: 51 KAKKTNQKRQREPRFAFMT-----------KSEVDHLEDGYRWRKYGQKAVKNSPFPRSY 197
Query: 103 YRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPL 141
YRC+S+ C +K+VERS DP+ ++ TY +H HS P+
Sbjct: 198 YRCTSA-SCNVKKRVERSFTDPSIVVTTYEGQHTHSSPV 311
>TC8298 homologue to UP|Q9SXP4 (Q9SXP4) DNA-binding protein NtWRKY3,
partial (16%)
Length = 514
Score = 62.0 bits (149), Expect(2) = 1e-14
Identities = 26/38 (68%), Positives = 30/38 (78%)
Frame = +3
Query: 100 RGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNH 137
RGYY+CSS KGCPARK VER++ DP L+VTY EH H
Sbjct: 159 RGYYKCSSEKGCPARKHVERAQDDPNMLIVTYEGEHRH 272
Score = 33.5 bits (75), Expect(2) = 1e-14
Identities = 13/13 (100%), Positives = 13/13 (100%)
Frame = +1
Query: 88 YGQKPIKGSPYPR 100
YGQKPIKGSPYPR
Sbjct: 1 YGQKPIKGSPYPR 39
>TC8329 similar to UP|BAD06717 (BAD06717) WRKY transcription factor 1,
partial (41%)
Length = 1717
Score = 74.7 bits (182), Expect = 1e-14
Identities = 44/136 (32%), Positives = 70/136 (51%)
Frame = +1
Query: 35 SPSSCEDTKIEESSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIK 94
S SS D E+S K R E K ++ E + + + D + WRKYGQK +
Sbjct: 466 SESSSTD---EDSFKKPREETIKAKISRAYVRTEATDTG---FVVKDGYQWRKYGQKVTR 627
Query: 95 GSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSHHSSTNAVTA 154
+P PR Y++CS + CP +K+V+RS D + L+ TY EHNH P + +S +
Sbjct: 628 DNPCPRAYFKCSFAPSCPVKKKVQRSVDDQSVLVATYEGEHNHPQP-SQMEATSGSGRNV 804
Query: 155 TVTAAVDTPSPVESPA 170
++ ++ + + SPA
Sbjct: 805 SLVGSMPSSKSLSSPA 852
>TC11734 similar to UP|WR49_ARATH (Q9FHR7) Probable WRKY transcription
factor 49 (WRKY DNA-binding protein 49), partial (28%)
Length = 1009
Score = 73.6 bits (179), Expect = 3e-14
Identities = 33/57 (57%), Positives = 40/57 (69%)
Frame = +3
Query: 81 DSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNH 137
D + WRKYGQK IK SP PR YYRC++ + C A+KQVERS DP L++TY H H
Sbjct: 336 DGYKWRKYGQKSIKNSPNPRSYYRCTNPR-CSAKKQVERSNEDPETLIITYEGLHLH 503
>AV427157
Length = 379
Score = 70.5 bits (171), Expect = 3e-13
Identities = 41/111 (36%), Positives = 59/111 (52%), Gaps = 4/111 (3%)
Frame = +3
Query: 31 DGPASPSSCEDTKIE----ESSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWR 86
DG S + K+E E S R + RVV ++V+ D + WR
Sbjct: 75 DGEGDESESKRRKLESYATELSGATRAIREPRVVVQTTSEVDIL---------DDGYRWR 227
Query: 87 KYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNH 137
KYGQK +KG+P PR YY+C+++ GC RK VER+ D +++ TY +HNH
Sbjct: 228 KYGQKVVKGNPNPRSYYKCTNA-GCMVRKHVERASHDLKSVITTYEGKHNH 377
>BP046076
Length = 593
Score = 66.6 bits (161), Expect = 4e-12
Identities = 39/110 (35%), Positives = 58/110 (52%)
Frame = -2
Query: 45 EESSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYR 104
E+S K RR++++ ++R + D + WRKYGQK +K S +PR YYR
Sbjct: 538 EKSKVKVRRKLREPRFCF--------QTRSDVDVLDDGYKWRKYGQKVVKNSLHPRSYYR 383
Query: 105 CSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSHHSSTNAVTA 154
C+ S C +K+VER D ++ TY HNHS P +S+ S T+
Sbjct: 382 CTHS-NCRVKKRVERLSEDCRMVITTYEGRHNHS-PCDESNSSEHECFTS 239
>TC11124 similar to UP|Q8H9D7 (Q8H9D7) WRKY-type DNA binding protein,
partial (45%)
Length = 517
Score = 57.0 bits (136), Expect = 3e-09
Identities = 25/59 (42%), Positives = 34/59 (57%)
Frame = +1
Query: 81 DSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSL 139
D + WRKYGQK +K + +PR YYRC + GC +K V R D ++ TY H H +
Sbjct: 1 DGYRWRKYGQKAVKNNKFPRSYYRC-THHGCHVKKPVPRLTQDEGVVVTTYEGVHTHPI 174
>TC16534 similar to UP|WR41_ARATH (Q8H0Y8) Probable WRKY transcription
factor 41 (WRKY DNA-binding protein 41), partial (20%)
Length = 697
Score = 53.1 bits (126), Expect = 5e-08
Identities = 25/66 (37%), Positives = 34/66 (50%)
Frame = +1
Query: 41 DTKIEESSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPR 100
D I++ KR K++ + I + G P D + WRKYGQK I G+ YPR
Sbjct: 469 DWAIQDHQEAKRDSKKRKTMPKWIDQIRVVSENGLEGPHDDGYNWRKYGQKDILGAKYPR 648
Query: 101 GYYRCS 106
YYRC+
Sbjct: 649 SYYRCT 666
>TC14985 similar to UP|WR65_ARATH (Q9LP56) Probable WRKY transcription
factor 65 (WRKY DNA-binding protein 65), partial (13%)
Length = 608
Score = 50.4 bits (119), Expect = 3e-07
Identities = 34/70 (48%), Positives = 38/70 (53%), Gaps = 11/70 (15%)
Frame = +1
Query: 188 DLTGDHHAVFGWFDDIVSTG-----VLVSPICG------GVEDVTLTMREEDESLFADLG 236
DL G++ GW + T +L SPI V V L MREEDE LFADLG
Sbjct: 7 DLGGENE--LGWLGVEMETASSAAAMLESPIMAPEFDEADVASVFLPMREEDELLFADLG 180
Query: 237 ELPECSTVFR 246
ELPECS VFR
Sbjct: 181 ELPECSAVFR 210
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.132 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,107,927
Number of Sequences: 28460
Number of extensions: 85290
Number of successful extensions: 756
Number of sequences better than 10.0: 115
Number of HSP's better than 10.0 without gapping: 686
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 731
length of query: 261
length of database: 4,897,600
effective HSP length: 88
effective length of query: 173
effective length of database: 2,393,120
effective search space: 414009760
effective search space used: 414009760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC138015.8


