

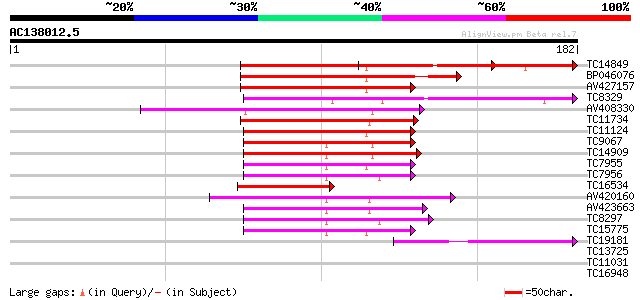
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138012.5 + phase: 0
(182 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14849 similar to UP|Q40090 (Q40090) SPF1 protein, partial (37%) 77 2e-15
BP046076 72 6e-14
AV427157 70 2e-13
TC8329 similar to UP|BAD06717 (BAD06717) WRKY transcription fact... 69 6e-13
AV408330 67 2e-12
TC11734 similar to UP|WR49_ARATH (Q9FHR7) Probable WRKY transcri... 65 5e-12
TC11124 similar to UP|Q8H9D7 (Q8H9D7) WRKY-type DNA binding prot... 61 1e-10
TC9067 similar to UP|WR15_ARATH (O22176) Probable WRKY transcrip... 53 3e-08
TC14909 similar to UP|WR21_ARATH (O04336) Probable WRKY transcri... 51 1e-07
TC7955 similar to UP|WR11_ARATH (Q9SV15) Probable WRKY transcrip... 50 3e-07
TC7956 similar to GB|AAM61419.1|21537078|AY084855 putaive DNA-bi... 49 7e-07
TC16534 similar to UP|WR41_ARATH (Q8H0Y8) Probable WRKY transcri... 48 1e-06
AV420160 47 3e-06
AV423663 46 3e-06
TC8297 similar to UP|Q40828 (Q40828) WRKY3, partial (27%) 46 3e-06
TC15775 similar to UP|WR17_ARATH (Q9SJA8) Probable WRKY transcri... 46 3e-06
TC19181 similar to UP|Q40090 (Q40090) SPF1 protein, partial (19%) 41 1e-04
TC13725 similar to UP|Q39658 (Q39658) SPF1-like DNA-binding prot... 34 0.013
TC11031 homologue to UP|WR65_ARATH (Q9LP56) Probable WRKY transc... 33 0.022
TC16948 28 1.2
>TC14849 similar to UP|Q40090 (Q40090) SPF1 protein, partial (37%)
Length = 1389
Score = 76.6 bits (187), Expect = 2e-15
Identities = 39/83 (46%), Positives = 51/83 (60%), Gaps = 1/83 (1%)
Frame = +2
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT P C V+K VER + D + T Y+G HNH P
Sbjct: 404 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVP 583
Query: 134 TSSMKRNSSSEYLYSLLPSETGS 156
+ R S + + LP+ T +
Sbjct: 584 AA---RGSGNHSVTRPLPNNTAN 643
Score = 61.6 bits (148), Expect = 8e-11
Identities = 35/73 (47%), Positives = 48/73 (64%), Gaps = 3/73 (4%)
Frame = +2
Query: 113 RTIDGEVIETLYKGTHNHWKPTSSMKRNSSSEYLYSLLPSETGSIDLQDQSF---GSEQL 169
R++DG++ E +YKGTHNH KP ++ +RNSSS + S S D+QDQS+ GS Q+
Sbjct: 2 RSLDGQITEIVYKGTHNHPKPQAT-RRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQM 178
Query: 170 DSDEEPTKSSVSI 182
DS P SS+SI
Sbjct: 179 DSVATPENSSISI 217
>BP046076
Length = 593
Score = 72.0 bits (175), Expect = 6e-14
Identities = 36/72 (50%), Positives = 48/72 (66%), Gaps = 1/72 (1%)
Frame = -2
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV K S + RSYY+CT NC VKK+VER + D ++ T Y+G HNH
Sbjct: 457 DDGYKWRKYGQKVVKNSLHPRSYYRCTHSNCRVKKRVERLSEDCRMVITTYEGRHNH--- 287
Query: 134 TSSMKRNSSSEY 145
S ++SSE+
Sbjct: 286 -SPCDESNSSEH 254
>AV427157
Length = 379
Score = 70.5 bits (171), Expect = 2e-13
Identities = 32/57 (56%), Positives = 40/57 (70%), Gaps = 1/57 (1%)
Frame = +3
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNH 130
DDGY W+KY +KV KG+ N RSYYKCT C V+K VER + D + + T Y+G HNH
Sbjct: 207 DDGYRWRKYGQKVVKGNPNPRSYYKCTNAGCMVRKHVERASHDLKSVITTYEGKHNH 377
>TC8329 similar to UP|BAD06717 (BAD06717) WRKY transcription factor 1,
partial (41%)
Length = 1717
Score = 68.6 bits (166), Expect = 6e-13
Identities = 43/119 (36%), Positives = 64/119 (53%), Gaps = 12/119 (10%)
Frame = +1
Query: 76 DGYNWKKYEEKVAKGSENQRSYYKCTW-PNCFVKKKVERTIDGE-VIETLYKGTHNHWKP 133
DGY W+KY +KV + + R+Y+KC++ P+C VKKKV+R++D + V+ Y+G HNH +P
Sbjct: 586 DGYQWRKYGQKVTRDNPCPRAYFKCSFAPSCPVKKKVQRSVDDQSVLVATYEGEHNHPQP 765
Query: 134 TSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGSEQLD----------SDEEPTKSSVSI 182
S M+ S S SL+ S S L + LD + EP K S +
Sbjct: 766 -SQMEATSGSGRNVSLVGSMPSSKSLSSPAPAVVTLDLTKSRCSNDSKNAEPRKDSAKL 939
>AV408330
Length = 316
Score = 67.0 bits (162), Expect = 2e-12
Identities = 34/98 (34%), Positives = 53/98 (53%), Gaps = 7/98 (7%)
Frame = +3
Query: 43 EIDGVDNNRNEFQSDYSNGRQQNKSSVRRMNH------DDGYNWKKYEEKVAKGSENQRS 96
E D + + + ++ +N ++Q + M +DGY W+KY +K K S RS
Sbjct: 15 EDDEHNKTKKQLKAKKTNQKRQREPRFAFMTKSEVDHLEDGYRWRKYGQKAVKNSPFPRS 194
Query: 97 YYKCTWPNCFVKKKVERTI-DGEVIETLYKGTHNHWKP 133
YY+CT +C VKK+VER+ D ++ T Y+G H H P
Sbjct: 195 YYRCTSASCNVKKRVERSFTDPSIVVTTYEGQHTHSSP 308
>TC11734 similar to UP|WR49_ARATH (Q9FHR7) Probable WRKY transcription
factor 49 (WRKY DNA-binding protein 49), partial (28%)
Length = 1009
Score = 65.5 bits (158), Expect = 5e-12
Identities = 30/58 (51%), Positives = 38/58 (64%), Gaps = 1/58 (1%)
Frame = +3
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERT-IDGEVIETLYKGTHNHW 131
DDGY W+KY +K K S N RSYY+CT P C KK+VER+ D E + Y+G H H+
Sbjct: 333 DDGYKWRKYGQKSIKNSPNPRSYYRCTNPRCSAKKQVERSNEDPETLIITYEGLHLHF 506
>TC11124 similar to UP|Q8H9D7 (Q8H9D7) WRKY-type DNA binding protein,
partial (45%)
Length = 517
Score = 61.2 bits (147), Expect = 1e-10
Identities = 28/56 (50%), Positives = 35/56 (62%), Gaps = 1/56 (1%)
Frame = +1
Query: 76 DGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNH 130
DGY W+KY +K K ++ RSYY+CT C VKK V R T D V+ T Y+G H H
Sbjct: 1 DGYRWRKYGQKAVKNNKFPRSYYRCTHHGCHVKKPVPRLTQDEGVVVTTYEGVHTH 168
>TC9067 similar to UP|WR15_ARATH (O22176) Probable WRKY transcription
factor 15 (WRKY DNA-binding protein 15), partial (31%)
Length = 660
Score = 53.1 bits (126), Expect = 3e-08
Identities = 25/57 (43%), Positives = 35/57 (60%), Gaps = 2/57 (3%)
Frame = +3
Query: 76 DGYNWKKYEEKVAKGSENQRSYYKC-TWPNCFVKKKVERTI-DGEVIETLYKGTHNH 130
D Y+W+KY +K KGS + R YYKC + C +K VER + D ++ Y+G HNH
Sbjct: 126 DDYSWRKYGQKPIKGSPHPRGYYKCSSVRGCPARKHVERALDDAAMLVVTYEGEHNH 296
>TC14909 similar to UP|WR21_ARATH (O04336) Probable WRKY transcription
factor 21 (WRKY DNA-binding protein 21), partial (22%)
Length = 590
Score = 51.2 bits (121), Expect = 1e-07
Identities = 25/59 (42%), Positives = 36/59 (60%), Gaps = 2/59 (3%)
Frame = +3
Query: 76 DGYNWKKYEEKVAKGSENQRSYYKC-TWPNCFVKKKVERTI-DGEVIETLYKGTHNHWK 132
D Y+W+KY +K KGS + R YYKC + C +K VER + + ++ Y+G HNH K
Sbjct: 63 DDYSWRKYGQKPIKGSPHPRGYYKCSSMRGCPARKHVERCLEEPTMLLVTYEGEHNHPK 239
>TC7955 similar to UP|WR11_ARATH (Q9SV15) Probable WRKY transcription
factor 11 (WRKY DNA-binding protein 11), partial (27%)
Length = 579
Score = 49.7 bits (117), Expect = 3e-07
Identities = 26/57 (45%), Positives = 33/57 (57%), Gaps = 2/57 (3%)
Frame = +3
Query: 76 DGYNWKKYEEKVAKGSENQRSYYKC-TWPNCFVKKKVER-TIDGEVIETLYKGTHNH 130
D Y+W+KY +K KGS R YYKC T C +K VER T D ++ Y+G H H
Sbjct: 93 DEYSWRKYGQKPIKGSPYPRGYYKCSTVRGCPARKHVERATDDPTMLIVTYEGEHRH 263
>TC7956 similar to GB|AAM61419.1|21537078|AY084855 putaive DNA-binding
protein {Arabidopsis thaliana;} , partial (45%)
Length = 1257
Score = 48.5 bits (114), Expect = 7e-07
Identities = 25/57 (43%), Positives = 32/57 (55%), Gaps = 2/57 (3%)
Frame = +3
Query: 76 DGYNWKKYEEKVAKGSENQRSYYKC-TWPNCFVKKKVERTIDG-EVIETLYKGTHNH 130
D Y+W+KY +K KGS R YYKC T C +K VER D ++ Y+G H H
Sbjct: 654 DEYSWRKYGQKPIKGSPYPRGYYKCSTVRGCPARKHVERAQDDPNMLIVTYEGEHRH 824
>TC16534 similar to UP|WR41_ARATH (Q8H0Y8) Probable WRKY transcription
factor 41 (WRKY DNA-binding protein 41), partial (20%)
Length = 697
Score = 47.8 bits (112), Expect = 1e-06
Identities = 18/31 (58%), Positives = 24/31 (77%)
Frame = +1
Query: 74 HDDGYNWKKYEEKVAKGSENQRSYYKCTWPN 104
HDDGYNW+KY +K G++ RSYY+CT+ N
Sbjct: 583 HDDGYNWRKYGQKDILGAKYPRSYYRCTFRN 675
>AV420160
Length = 417
Score = 46.6 bits (109), Expect = 3e-06
Identities = 26/81 (32%), Positives = 40/81 (49%), Gaps = 2/81 (2%)
Frame = +1
Query: 65 NKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKC-TWPNCFVKKKVERT-IDGEVIET 122
+KS D + W+KY +K KGS R YY+C + C +K+VER+ +D +
Sbjct: 124 SKSKGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTKLMV 303
Query: 123 LYKGTHNHWKPTSSMKRNSSS 143
Y HNH P +++S
Sbjct: 304 TYSYEHNHSLPVPKSHSSAAS 366
>AV423663
Length = 447
Score = 46.2 bits (108), Expect = 3e-06
Identities = 24/61 (39%), Positives = 33/61 (53%), Gaps = 2/61 (3%)
Frame = +1
Query: 76 DGYNWKKYEEKVAKGSENQRSYYKC-TWPNCFVKKKVERT-IDGEVIETLYKGTHNHWKP 133
D + W+KY +K KGS R YY+C + C +K+VER + + Y G HNH P
Sbjct: 82 DIWAWRKYGQKPIKGSPYPRGYYRCSSSKGCLARKQVERNKSEPAMFIVTYTGEHNHPAP 261
Query: 134 T 134
T
Sbjct: 262 T 264
>TC8297 similar to UP|Q40828 (Q40828) WRKY3, partial (27%)
Length = 617
Score = 46.2 bits (108), Expect = 3e-06
Identities = 24/63 (38%), Positives = 34/63 (53%), Gaps = 2/63 (3%)
Frame = +1
Query: 76 DGYNWKKYEEKVAKGSENQRSYYKC-TWPNCFVKKKVERTIDG-EVIETLYKGTHNHWKP 133
D + W+KY +K KGS R YYKC + C +K VER D ++ Y+G H H
Sbjct: 172 DDFQWRKYGQKPIKGSPYPRGYYKCSSEKGCPARKHVERAQDDPNMLIVTYEGEHRHLIT 351
Query: 134 TSS 136
T++
Sbjct: 352 TTA 360
>TC15775 similar to UP|WR17_ARATH (Q9SJA8) Probable WRKY transcription
factor 17 (WRKY DNA-binding protein 17), partial (53%)
Length = 1284
Score = 46.2 bits (108), Expect = 3e-06
Identities = 24/57 (42%), Positives = 33/57 (57%), Gaps = 2/57 (3%)
Frame = +3
Query: 76 DGYNWKKYEEKVAKGSENQRSYYKC-TWPNCFVKKKVER-TIDGEVIETLYKGTHNH 130
D Y+W+KY +K KG+ R YYKC T C +K VER T D ++ Y+ H+H
Sbjct: 741 DEYSWRKYGQKPIKGTPYPRGYYKCSTVRGCPARKHVERATDDPAMLIVTYEDEHDH 911
>TC19181 similar to UP|Q40090 (Q40090) SPF1 protein, partial (19%)
Length = 402
Score = 41.2 bits (95), Expect = 1e-04
Identities = 25/59 (42%), Positives = 32/59 (53%)
Frame = +3
Query: 124 YKGTHNHWKPTSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGSEQLDSDEEPTKSSVSI 182
YKGTHNH KP + KRNS S S S ++Q + S Q+DS P SS+S+
Sbjct: 3 YKGTHNHPKPQAPHKRNS------SFTTSNHVSTEIQGATHVSGQMDSVATPENSSLSM 161
Score = 33.9 bits (76), Expect = 0.017
Identities = 12/19 (63%), Positives = 15/19 (78%)
Frame = +3
Query: 75 DDGYNWKKYEEKVAKGSEN 93
DDGY W+KY +KV KG+ N
Sbjct: 345 DDGYRWRKYGQKVVKGNPN 401
>TC13725 similar to UP|Q39658 (Q39658) SPF1-like DNA-binding protein,
partial (11%)
Length = 578
Score = 34.3 bits (77), Expect = 0.013
Identities = 24/76 (31%), Positives = 38/76 (49%), Gaps = 5/76 (6%)
Frame = +2
Query: 105 CFVKKKVER-TIDGEVIETLYKGTHNHWKPTS-SMKRNSSSEYLYSLLPSETGSID---L 159
C V+K VER + D + + T Y+G HNH P + + N++S P G L
Sbjct: 5 CNVRKHVERASTDAKAVITTYEGKHNHDVPAARNSSHNTASSNSMQPKPQNMGPEKHPLL 184
Query: 160 QDQSFGSEQLDSDEEP 175
+D FG+ ++D+ P
Sbjct: 185 KDMDFGNN--NNDQRP 226
>TC11031 homologue to UP|WR65_ARATH (Q9LP56) Probable WRKY transcription
factor 65 (WRKY DNA-binding protein 65), partial (15%)
Length = 453
Score = 33.5 bits (75), Expect = 0.022
Identities = 12/26 (46%), Positives = 17/26 (65%)
Frame = +3
Query: 76 DGYNWKKYEEKVAKGSENQRSYYKCT 101
D + W+KY +K KGS R YY+C+
Sbjct: 351 DSWAWRKYGQKPIKGSPYPRGYYRCS 428
>TC16948
Length = 460
Score = 27.7 bits (60), Expect = 1.2
Identities = 15/47 (31%), Positives = 23/47 (48%), Gaps = 1/47 (2%)
Frame = +1
Query: 93 NQRSYYKCTWPNCFVKKKVERTIDG-EVIETLYKGTHNHWKPTSSMK 138
+QRSY TW C++ K + T G ++ + K H K +S K
Sbjct: 25 SQRSYILMTWR*CYLMKDPQATAHGLQINDAALKVLHRQRKEEASYK 165
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.311 0.129 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,046,406
Number of Sequences: 28460
Number of extensions: 38728
Number of successful extensions: 213
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 204
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 206
length of query: 182
length of database: 4,897,600
effective HSP length: 85
effective length of query: 97
effective length of database: 2,478,500
effective search space: 240414500
effective search space used: 240414500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 52 (24.6 bits)
Medicago: description of AC138012.5


