

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137995.11 + phase: 0
(1094 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
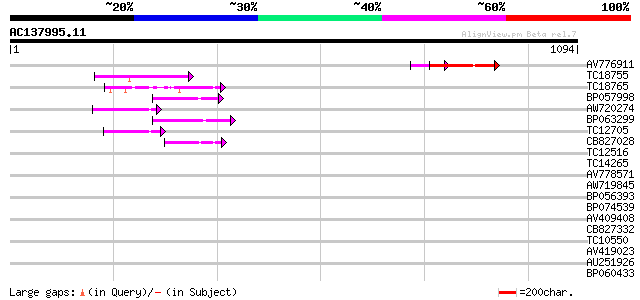
Score E
Sequences producing significant alignments: (bits) Value
AV776911 103 1e-22
TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fr... 76 3e-14
TC18765 weakly similar to UP|Q9LVT1 (Q9LVT1) Disease resistance ... 63 3e-10
BP057998 59 5e-09
AW720274 56 4e-08
BP063299 49 6e-06
TC12705 48 1e-05
CB827028 43 2e-04
TC12516 similar to UP|Q9M5V7 (Q9M5V7) Disease resistance-like pr... 39 0.006
TC14265 weakly similar to UP|AAN62760 (AAN62760) Disease resista... 38 0.008
AV778571 37 0.017
AW719845 36 0.029
BP056393 35 0.064
BP074539 32 0.42
AV409408 32 0.54
CB827332 32 0.71
TC10550 weakly similar to UP|Q40392 (Q40392) Virus resistance (N... 32 0.71
AV419023 31 0.93
AU251926 31 0.93
BP060433 30 1.6
>AV776911
Length = 432
Score = 103 bits (257), Expect = 1e-22
Identities = 60/138 (43%), Positives = 85/138 (61%), Gaps = 3/138 (2%)
Frame = +1
Query: 810 QLEKLQVLSCPELQHILIDDDRDEI---SAYDYRLLLFPKLKKFHVRECGVLEYIIPITL 866
QLE L++ C L+HI+ +D + + + +F KLK+ V+ CG LE + P++
Sbjct: 4 QLEVLKIRHCDGLKHIMTGEDAEIVLDDNGQQVFRSVFTKLKRLIVKWCGQLESLFPVSF 183
Query: 867 AQGLVQLECLEIVCNENLKYVFGQSTHNDGQNQNELKIIELSALEELTLVNLPNINSICP 926
++GLVQLE LEI LKY+F Q+ + QNQNEL IIE L+ L L++LPNI +ICP
Sbjct: 184 SKGLVQLEYLEIDKAPELKYIFYQNQLSC-QNQNELHIIEFPVLKLLKLLHLPNIINICP 360
Query: 927 EDCYLMWPSLLQFNLQNC 944
E+ + MWPSL L C
Sbjct: 361 ENYHPMWPSLQNLYLLQC 414
Score = 43.1 bits (100), Expect = 2e-04
Identities = 23/76 (30%), Positives = 39/76 (51%)
Frame = +1
Query: 773 NGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDDDRD 832
NGQ F L+ L + C +L LF ++ ++ L QLE L++ PEL++I +
Sbjct: 88 NGQQVFRSVFTKLKRLIVKWCGQLESLFPVSFSKGLVQLEYLEIDKAPELKYIFYQNQLS 267
Query: 833 EISAYDYRLLLFPKLK 848
+ + ++ FP LK
Sbjct: 268 CQNQNELHIIEFPVLK 315
>TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fragment),
complete
Length = 880
Score = 76.3 bits (186), Expect = 3e-14
Identities = 56/204 (27%), Positives = 95/204 (46%), Gaps = 13/204 (6%)
Frame = +3
Query: 164 CALKDDDVT-MIGLYGMGGCGKTMLAMEVGKRCGNL--FDQVLFVPISSTVEVERIQEKI 220
C +K V +I + GMGG GKT L +V + F ++ +S + E+ + +
Sbjct: 234 CLIKPCPVRKVISVTGMGGMGKTTLVKQVYDDPVVIKHFRACAWITVSQSCEIGELLRDL 413
Query: 221 AGSLEFEFQ-------EKDEMDRSKRLCMRLTQEDRVLVILDDVWQMLDFDAIGIPSIEH 273
A L E + E DR K + L Q R LV+ DDVW + +++A+ ++
Sbjct: 414 ARQLFSEIRRPVPLGLENMRCDRLKMIIKDLLQRRRYLVVFDDVWHVREWEAVKYALPDN 593
Query: 274 HKGCKILITSRSEAVCTLMDCQKK---IQLSTLTNDETWDLFQKQALISEGTWISIKNMA 330
+ G +I+IT+R + + K L L DE W+LF ++ + + +
Sbjct: 594 NCGSRIMITTRRSDLAFTSSTESKGKVYNLQPLKEDEAWELFCRKTFHGDSCPSHLIGIC 773
Query: 331 REISNECKGLPVATVAVASSLKGK 354
I +C+GLP+A VA++ L K
Sbjct: 774 TYILRKCEGLPLAIVAISGVLATK 845
>TC18765 weakly similar to UP|Q9LVT1 (Q9LVT1) Disease resistance
protein-like, partial (18%)
Length = 688
Score = 62.8 bits (151), Expect = 3e-10
Identities = 69/261 (26%), Positives = 115/261 (43%), Gaps = 28/261 (10%)
Frame = +2
Query: 184 KTMLAMEVGK----RCGNLFDQVLFVPISSTVEVERIQEKIAG------SLEFEFQEKDE 233
KT LA EV + RC + +++LF+ +S + VE ++ KI G L +
Sbjct: 2 KTTLAREVCRDDQVRC-HFKERILFLTVSQSPNVEELRAKIFGHIMGNRGLNANYAVPQW 178
Query: 234 MDRSKRLCMRLTQEDRVLVILDDVWQM--LDFDAIGIPSIEHHKGCKILITSRSEAVCTL 291
M + + ++LV+LDDVW + L+ + +P GCK L+ SR
Sbjct: 179 MPQ-----FECQSQSQILVVLDDVWSLPVLEQLVLRVP------GCKYLVVSR------- 304
Query: 292 MDCQKKIQLSTLTNDETWDLFQKQALISEGTWISI----------------KNMAREISN 335
+ + ND T+D+ L+SEG +S+ +N+ +++
Sbjct: 305 ------FKFQRIFND-TYDV----ELLSEGDALSLFCHHAFGHKSIPFGANQNLIKQVVA 451
Query: 336 ECKGLPVATVAVASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTE 395
EC LP+A + +SL+ + E+ W RL + E N + +S + L E
Sbjct: 452 ECGRLPLALKVIGASLRDQNEMFWLSVKTRLSQGLSIG-ESYEVNLIDRMAISTNYLP-E 625
Query: 396 EAKSLFLLCSVFPEDCEIPVE 416
+ K FL FPED +IP+E
Sbjct: 626 KVKECFLDLCAFPEDKKIPLE 688
>BP057998
Length = 566
Score = 58.5 bits (140), Expect = 5e-09
Identities = 39/142 (27%), Positives = 72/142 (50%), Gaps = 4/142 (2%)
Frame = +2
Query: 275 KGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQALISEG---TWISIKNMAR 331
+G KIL+T+RS+ V +++ + LS L+N++ W +F A + G I+++ +
Sbjct: 62 RGSKILVTTRSDEVASVVQTGQTFNLSQLSNEDCWSVFANHACLFPGLGENTIALEKIGL 241
Query: 332 EISNECKGLPVATVAVASSLKGKAEV-EWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYD 390
EI +CKGLP+A ++ L+ K + +W L + + + L++SY
Sbjct: 242 EIVKKCKGLPLAAQSLGGLLRRKHNIKDWNNVL----NCDIWELSESESKIIPSLRISYH 409
Query: 391 NLDTEEAKSLFLLCSVFPEDCE 412
L + K F S++P+D E
Sbjct: 410 YLPS-YLKRCFAYFSLYPKDYE 472
>AW720274
Length = 523
Score = 55.8 bits (133), Expect = 4e-08
Identities = 42/136 (30%), Positives = 69/136 (49%), Gaps = 4/136 (2%)
Frame = +2
Query: 161 ELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKRCGNLFDQVLFVPISSTVEVERIQEKI 220
+L+ AL+DD IGL+G G GKT L +FD V+FV + +++ ++Q +I
Sbjct: 98 DLLNALQDDTCHSIGLFGERGSGKTALVKAEIPEYRKIFDAVVFVTV-KPLDIWKLQHQI 274
Query: 221 AGSLEFEFQE-KDEMDRSKRLCMRLTQEDRVLVILDDV-WQMLDFDA--IGIPSIEHHKG 276
A + K + R+ + + +++ LVI DDV DF+ IGIP+
Sbjct: 275 ALQFRINMESVKKQSARASVIHSAIKSKNKALVIFDDVSTGTADFNLQNIGIPN--PSMK 448
Query: 277 CKILITSRSEAVCTLM 292
CKIL+ + + C +M
Sbjct: 449 CKILLITHDKNFCNMM 496
>BP063299
Length = 580
Score = 48.5 bits (114), Expect = 6e-06
Identities = 43/164 (26%), Positives = 75/164 (45%), Gaps = 4/164 (2%)
Frame = +3
Query: 276 GC--KILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQAL-ISEGTWISIKNMARE 332
GC +L+++R V +M + +L L+ ++ W LF++ A + + + +E
Sbjct: 39 GCTQSVLVSTRDMDVAEIMGTCQAQRLQGLSENDCWLLFKQYAFGANNEERAELVAIGKE 218
Query: 333 ISNECKGLPVATVAVASSLKGKAEV-EWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDN 391
I +C GLP+A A+ L+ K EV EW + + P + L LSY +
Sbjct: 219 IVKKCGGLPLAARALGGLLRLKNEVKEWHEVKESTLWNLPDE-----RCILPALMLSYFH 383
Query: 392 LDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVGEVHSYE 435
L T + F C++FP+D +I E L + G + + E
Sbjct: 384 L-TPTLRQCFAFCAIFPKDAKIIKEDLIYLWMANGFISSTKNKE 512
>TC12705
Length = 376
Score = 47.8 bits (112), Expect = 1e-05
Identities = 37/125 (29%), Positives = 61/125 (48%), Gaps = 5/125 (4%)
Frame = +2
Query: 181 GCGKTMLAMEVGKRCGNLFDQVLFVPISSTVEVERIQEKIAGSLEFEFQE-KDEMDRSKR 239
G GKT L N+F V++V +S ++ ++Q +IA L + + K + R+
Sbjct: 8 GSGKTALVKAEIPEYENIFHLVVYVTVSEQADIAKLQNQIAHQLNLKLKAVKSKHRRASA 187
Query: 240 LCMRLTQE-DRVLVILDDV---WQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQ 295
+ L + +VLVI DDV + D + IGIP + K KIL+ + + C ++ C
Sbjct: 188 IESALLKSVGKVLVIFDDVSTNFGPQDIEDIGIP--DPSKQLKILLITHDKKYCNMIRCN 361
Query: 296 KKIQL 300
I L
Sbjct: 362 PWIPL 376
>CB827028
Length = 572
Score = 43.1 bits (100), Expect = 2e-04
Identities = 37/119 (31%), Positives = 51/119 (42%)
Frame = +2
Query: 300 LSTLTNDETWDLFQKQALISEGTWISIKNMAREISNECKGLPVATVAVASSLKGKAEVEW 359
L L+ D LF A +S + ++ ++I CK P+A +SLK K W
Sbjct: 47 LERLSLDYATSLFLHSASLSPNSTEIPVSLVKKIVKGCKRSPIALTVTGTSLKNKNLAVW 226
Query: 360 KVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFL 418
+ L SK +I + + CLQ S D LD A F +FPED IP L
Sbjct: 227 RSRAKAL--SKGQSILESNHDVLTCLQKSLDVLD-PIAIECFKDLGLFPEDQRIPAAAL 394
>TC12516 similar to UP|Q9M5V7 (Q9M5V7) Disease resistance-like protein
(Fragment), partial (36%)
Length = 859
Score = 38.5 bits (88), Expect = 0.006
Identities = 18/46 (39%), Positives = 30/46 (65%)
Frame = +3
Query: 160 EELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKRCGNLFDQVLFV 205
E L+C + DV +IG++GMGG GKT +A E+ R + ++ + F+
Sbjct: 579 ESLLCQ-ESKDVRVIGIWGMGGIGKTTIAEEIFNRRHSQYEGICFL 713
>TC14265 weakly similar to UP|AAN62760 (AAN62760) Disease resistance
protein-like protein MsR1, partial (5%)
Length = 766
Score = 38.1 bits (87), Expect = 0.008
Identities = 21/63 (33%), Positives = 35/63 (55%), Gaps = 1/63 (1%)
Frame = +1
Query: 578 VGDMKKLESITLCDCSFVE-LPDVVTQLTNLRLLDLSECGMERNPFEVIARHTELEELFF 636
+G ++ LE + L C+ ++ LPD + +L+ LRLLD+S C + E I L+ L+
Sbjct: 7 IGKLENLELLRLSSCTDLKGLPDSIGRLSKLRLLDISNCISLPSLPEEIGNLCNLKSLYM 186
Query: 637 ADC 639
C
Sbjct: 187 TSC 195
>AV778571
Length = 452
Score = 37.0 bits (84), Expect = 0.017
Identities = 45/144 (31%), Positives = 62/144 (42%), Gaps = 11/144 (7%)
Frame = +3
Query: 502 CEKFPNSLDCSNLDFLQIHTYTQVSDEIFK------GMRMLRVLFLYN--KGRERRPLLT 553
CE D S L L+ ++ S+ + K + LR+L + K R P+
Sbjct: 45 CESITQIPDLSGLPNLEKLSFVY-SENLIKIHESVGFLNKLRILDIEGCCKIRTFPPMKL 221
Query: 554 TSLKSLTNLRCILFSKWDLVDISFVGDMKKLESITLCDCSFV---ELPDVVTQLTNLRLL 610
SL+ L C SF + K+E+ITL ELP + LT LR L
Sbjct: 222 ASLEELYLSHCSSLE-------SFPEILAKMENITLIGVYHTPIKELPSSIGNLTRLRRL 380
Query: 611 DLSECGMERNPFEVIARHTELEEL 634
+L CGM + P +A TELEEL
Sbjct: 381 ELHGCGMLQLP-SGMAM*TELEEL 449
>AW719845
Length = 209
Score = 36.2 bits (82), Expect = 0.029
Identities = 22/63 (34%), Positives = 34/63 (53%), Gaps = 3/63 (4%)
Frame = +3
Query: 161 ELMCALKDDDVTMIGLYGMGGCGKTMLAMEV---GKRCGNLFDQVLFVPISSTVEVERIQ 217
+L L D V+++ + G+GG GKT LA V + G + +LFV S T V+ I
Sbjct: 9 KLKMELLKDGVSVVAVTGLGGLGKTTLAAMVCWDKQIKGKFRENILFVTFSKTPNVKNIV 188
Query: 218 EKI 220
E++
Sbjct: 189 ERL 197
>BP056393
Length = 421
Score = 35.0 bits (79), Expect = 0.064
Identities = 21/87 (24%), Positives = 42/87 (48%)
Frame = +1
Query: 324 ISIKNMAREISNECKGLPVATVAVASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPYK 383
++ + + R+I CKG P+ ++ L+GK ++ + + +S + + N
Sbjct: 106 MTFETIGRQIVKRCKGSPLXAQSLGGLLRGKHDIR---DXNNILNSNIWELPENESNIIL 276
Query: 384 CLQLSYDNLDTEEAKSLFLLCSVFPED 410
L++SY L K + CS+FP+D
Sbjct: 277 ALRISYHYL-PPHLKRCXVYCSLFPKD 354
>BP074539
Length = 357
Score = 32.3 bits (72), Expect = 0.42
Identities = 26/112 (23%), Positives = 54/112 (48%), Gaps = 2/112 (1%)
Frame = +1
Query: 554 TSLKSLTNLRCILFSKWDLVDISFVGDMKKLESITLCDCSFV-ELPDVVTQLTNLRLLDL 612
T+++ L + C+L++ L+ + L +C ++ ELP + +LTNLR LD
Sbjct: 37 TNIEKLPDSTCLLYN---------------LQILKLKNCRYLKELPLNLHKLTNLRYLDF 171
Query: 613 SECGMERNPFEV-IARHTELEELFFADCRSKWEVEFLKEFSVPQVLQRYQIQ 663
S + + P ++ ++ FF S++ ++ L E ++ L ++Q
Sbjct: 172 SGTQVRKMPMHFGKLKNLQVLNSFFVGQGSEYNIQQLDELNLQGTLSISELQ 327
>AV409408
Length = 430
Score = 32.0 bits (71), Expect = 0.54
Identities = 36/134 (26%), Positives = 59/134 (43%)
Frame = +2
Query: 339 GLPVATVAVASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAK 398
GLP+A + S L G++ +W+ AL +++ +I L++SYD L+ +E K
Sbjct: 5 GLPLALEVLGSFLCGRSLSDWEDALIKIKKVPHDDI-------LNKLRISYDMLE-DEHK 160
Query: 399 SLFLLCSVFPEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVN 458
+LFL + F + + GL TV N LI LL
Sbjct: 161 TLFLDIACFFKGWYKHKVIQLLGSCGL--------------HPTVGINVLIEKSLL--TF 292
Query: 459 EGKCVKMHDLVRNV 472
+G+ + MHDL+ +
Sbjct: 293 DGRVIGMHDLLEEM 334
>CB827332
Length = 511
Score = 31.6 bits (70), Expect = 0.71
Identities = 17/46 (36%), Positives = 25/46 (53%)
Frame = +3
Query: 784 NLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDD 829
NL+ L +S+CPK+T + A LE L V SCP + +D+
Sbjct: 297 NLKILKVSNCPKITVTGIGILVGKCACLEYLDVRSCPHITKAGLDE 434
>TC10550 weakly similar to UP|Q40392 (Q40392) Virus resistance (N), partial
(5%)
Length = 670
Score = 31.6 bits (70), Expect = 0.71
Identities = 16/40 (40%), Positives = 21/40 (52%)
Frame = +3
Query: 576 SFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSEC 615
S G +KLE + L LP + LT LR L+LS+C
Sbjct: 63 SSFGSQRKLEILDLEQGRIESLPSCIVNLTRLRYLNLSDC 182
>AV419023
Length = 410
Score = 31.2 bits (69), Expect = 0.93
Identities = 15/38 (39%), Positives = 23/38 (60%)
Frame = +3
Query: 578 VGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSEC 615
+G + LE + L +FVELP +++L L L+LS C
Sbjct: 54 IGRLWCLERLNLQGNNFVELPSTISRLRRLAYLNLSHC 167
>AU251926
Length = 335
Score = 31.2 bits (69), Expect = 0.93
Identities = 31/100 (31%), Positives = 50/100 (50%), Gaps = 1/100 (1%)
Frame = +2
Query: 514 LDFLQIHTYTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWDLV 573
L +++ + +VS + KG R L+ L NK ++ L SL L++L + S+ +V
Sbjct: 8 LSLIKLASLIEVSAK--KGTRDLK---LQNKLLDQVDWLPDSLGKLSSLVTLDLSENRIV 172
Query: 574 DI-SFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDL 612
+ S +G + L + L ELPD + L NL LDL
Sbjct: 173 ALPSTIGGLSSLTRLDLHTNRIQELPDSIGNLLNLVYLDL 292
>BP060433
Length = 365
Score = 30.4 bits (67), Expect = 1.6
Identities = 19/67 (28%), Positives = 31/67 (45%), Gaps = 8/67 (11%)
Frame = -3
Query: 964 INEASHQTLQNITEVRVNNCELEGIFQLVGLTNDGE--------KDPLTSCLEMLYLENL 1015
+N + ++ + +N+C + VG T+ GE K P L++LYLE+
Sbjct: 279 VNASLGAGIKAVKYEHINHC*IA*TIIYVGFTHTGEVMTLRGPVKQPCHRELQLLYLESS 100
Query: 1016 PQLRYLC 1022
P L Y C
Sbjct: 99 PTLWYSC 79
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,221,128
Number of Sequences: 28460
Number of extensions: 289319
Number of successful extensions: 1384
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 1360
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1379
length of query: 1094
length of database: 4,897,600
effective HSP length: 100
effective length of query: 994
effective length of database: 2,051,600
effective search space: 2039290400
effective search space used: 2039290400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137995.11


