

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137994.10 - phase: 0 /pseudo
(561 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
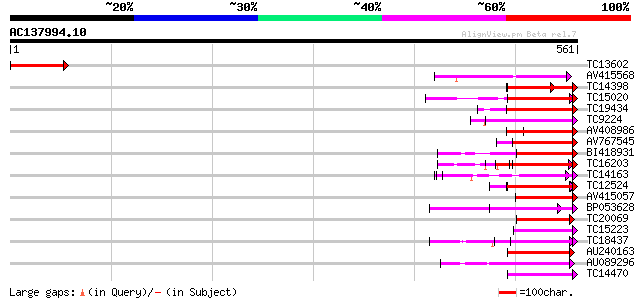
Score E
Sequences producing significant alignments: (bits) Value
TC13602 weakly similar to UP|Q84QD8 (Q84QD8) Avr9/Cf-9 rapidly e... 79 2e-15
AV415568 70 7e-13
TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%) 67 1e-11
TC15020 homologue to UP|Q9QX99 (Q9QX99) Transcription factor (T-... 64 6e-11
TC19434 similar to UP|Q94F63 (Q94F63) Somatic embryogenesis rece... 63 1e-10
TC9224 similar to GB|AAP21230.1|30102624|BT006422 At3g49750 {Ara... 62 3e-10
AV408986 60 9e-10
AV767545 60 9e-10
BI418931 59 2e-09
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 57 6e-09
TC14163 57 1e-08
TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kina... 55 4e-08
AV415057 53 1e-07
BP053628 53 1e-07
TC20069 similar to UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kina... 52 2e-07
TC15223 similar to UP|CAE76632 (CAE76632) Leucine rich repeat pr... 52 3e-07
TC18437 weakly similar to UP|Q9LVB3 (Q9LVB3) Receptor-like prote... 52 3e-07
AU240163 52 3e-07
AU089296 51 6e-07
TC14470 similar to UP|AAQ56728 (AAQ56728) Polygalacturonase inhi... 50 7e-07
>TC13602 weakly similar to UP|Q84QD8 (Q84QD8) Avr9/Cf-9 rapidly elicited
protein 275 (Fragment), partial (26%)
Length = 556
Score = 79.0 bits (193), Expect = 2e-15
Identities = 39/58 (67%), Positives = 46/58 (79%)
Frame = +3
Query: 1 HTNQVIHIDLSSSQLYGTLVANSSLFHLVHLQVLDLSDNDFNYSKIPSKIAYFLEKFH 58
HT VI IDLSSSQLYG L +NSSLF+L LQ+LDL+DNDFNYS+IPS+I F + H
Sbjct: 270 HTGHVIGIDLSSSQLYGYLDSNSSLFNLAQLQILDLADNDFNYSQIPSRIGEFSKLTH 443
Score = 36.6 bits (83), Expect = 0.011
Identities = 25/67 (37%), Positives = 42/67 (62%), Gaps = 3/67 (4%)
Frame = +3
Query: 498 NLIAIDISSNKISGEIPQ--VIEDLKGLVLLNLSNNLLTGS-IPSSLGKLINLEALDLSL 554
++I ID+SS+++ G + + +L L +L+L++N S IPS +G+ L L+LSL
Sbjct: 279 HVIGIDLSSSQLYGYLDSNSSLFNLAQLQILDLADNDFNYSQIPSRIGEFSKLTHLNLSL 458
Query: 555 NSLSGKV 561
S SG+V
Sbjct: 459 TSFSGEV 479
Score = 30.4 bits (67), Expect = 0.79
Identities = 20/61 (32%), Positives = 30/61 (48%), Gaps = 1/61 (1%)
Frame = +3
Query: 493 LQNLYNLIAIDISSNKIS-GEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALD 551
L NL L +D++ N + +IP I + L LNLS +G +P + L +LD
Sbjct: 342 LFNLAQLQILDLADNDFNYSQIPSRIGEFSKLTHLNLSLTSFSGEVPQEVSH*SKLLSLD 521
Query: 552 L 552
L
Sbjct: 522 L 524
>AV415568
Length = 412
Score = 70.5 bits (171), Expect = 7e-13
Identities = 46/138 (33%), Positives = 74/138 (53%), Gaps = 2/138 (1%)
Frame = +3
Query: 421 GRFTELKVLSLSNNEFHGDI--RCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESN 478
G + L+++ L+ N F G + +C + S + D + +Q + + +
Sbjct: 3 GTWKRLQIVDLAFNNFSGKLPGKCFTRWEAMMSGENQADSKVNHIRFQVLQYDQIYYQDS 182
Query: 479 EYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIP 538
+++KG +K+ ++ +ID SSN GEIP+ + D K L +LNLSNN L+G I
Sbjct: 183 VTVTSKGQGMELVKILTVFT--SIDFSSNHFQGEIPKELFDFKELYVLNLSNNALSGQIQ 356
Query: 539 SSLGKLINLEALDLSLNS 556
SS+G L LE+LDLS NS
Sbjct: 357 SSIGNLKQLESLDLSNNS 410
>TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%)
Length = 982
Score = 66.6 bits (161), Expect = 1e-11
Identities = 34/70 (48%), Positives = 47/70 (66%)
Frame = +1
Query: 492 KLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALD 551
+L NL +LI++D+ N +SG IP + +LK L L L+NN LTG IP SL L NL+ LD
Sbjct: 451 ELGNLQSLISLDLYHNNVSGSIPSSLGNLKNLRFLRLNNNHLTGQIPKSLSTLPNLKVLD 630
Query: 552 LSLNSLSGKV 561
+S N+L G +
Sbjct: 631 VSNNNLCGPI 660
Score = 58.5 bits (140), Expect = 3e-09
Identities = 30/69 (43%), Positives = 48/69 (69%)
Frame = +1
Query: 493 LQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDL 552
L NL++L +++ N I G IP+ + +L+ L+ L+L +N ++GSIPSSLG L NL L L
Sbjct: 382 LGNLHSLQYLELYENNIQGTIPEELGNLQSLISLDLYHNNVSGSIPSSLGNLKNLRFLRL 561
Query: 553 SLNSLSGKV 561
+ N L+G++
Sbjct: 562 NNNHLTGQI 588
Score = 41.2 bits (95), Expect = 4e-04
Identities = 20/48 (41%), Positives = 33/48 (68%)
Frame = +1
Query: 493 LQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSS 540
L NL NL + +++N ++G+IP+ + L L +L++SNN L G IP+S
Sbjct: 526 LGNLKNLRFLRLNNNHLTGQIPKSLSTLPNLKVLDVSNNNLCGPIPTS 669
>TC15020 homologue to UP|Q9QX99 (Q9QX99) Transcription factor (T-cell
leukemia, homeobox 1), partial (5%)
Length = 1304
Score = 63.9 bits (154), Expect = 6e-11
Identities = 32/69 (46%), Positives = 44/69 (63%)
Frame = +1
Query: 493 LQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDL 552
L +L NL + + SN SG IP I LK L L+ S+N L+GS+P+S+ LINL +DL
Sbjct: 361 LSSLSNLKTLTLRSNSFSGSIPPSITTLKSLYSLDFSHNSLSGSLPNSMNSLINLRRIDL 540
Query: 553 SLNSLSGKV 561
S N L+G +
Sbjct: 541 SFNKLTGSI 567
Score = 54.7 bits (130), Expect = 4e-08
Identities = 28/69 (40%), Positives = 40/69 (57%)
Frame = +1
Query: 493 LQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDL 552
+ L L+ +D++ N SG IP + L L L L +N +GSIP S+ L +L +LD
Sbjct: 289 ISQLTQLVTLDLAENSFSGPIPSSLSSLSNLKTLTLRSNSFSGSIPPSITTLKSLYSLDF 468
Query: 553 SLNSLSGKV 561
S NSLSG +
Sbjct: 469 SHNSLSGSL 495
Score = 48.5 bits (114), Expect = 3e-06
Identities = 43/148 (29%), Positives = 62/148 (41%)
Frame = +1
Query: 412 YQ*FISILAGRFTELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRN 471
Y ++ L + T+L L L+ N F G I S L SN
Sbjct: 265 YSGSLTPLISQLTQLVTLDLAENSFSGPIPSS--------------------LSSLSNLK 384
Query: 472 DPKLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNN 531
L SN + + + + L +L ++D S N +SG +P + L L ++LS N
Sbjct: 385 TLTLRSNSFSGS-----IPPSITTLKSLYSLDFSHNSLSGSLPNSMNSLINLRRIDLSFN 549
Query: 532 LLTGSIPSSLGKLINLEALDLSLNSLSG 559
LTGSIP L+ L + NSLSG
Sbjct: 550 KLTGSIPKLPPNLLELA---VKANSLSG 624
>TC19434 similar to UP|Q94F63 (Q94F63) Somatic embryogenesis receptor-like
kinase 2, partial (26%)
Length = 550
Score = 62.8 bits (151), Expect = 1e-10
Identities = 38/98 (38%), Positives = 56/98 (56%)
Frame = +2
Query: 464 LWQFSNRNDPKLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGL 523
L Q SN L+ E SN ++ +L NL NL+++D+ N ++G IP + +L L
Sbjct: 32 LGQLSN-----LQYLELYSNNMTGKIPDELGNLTNLVSLDLYLNNLTGNIPSTLGNLGKL 196
Query: 524 VLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
L L+NN L+G IP +L + +L+ LDLS N L G V
Sbjct: 197 RFLRLNNNTLSGGIPMTLTNVSSLQVLDLSNNQLKGVV 310
Score = 54.3 bits (129), Expect = 5e-08
Identities = 31/70 (44%), Positives = 46/70 (65%)
Frame = +2
Query: 492 KLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALD 551
+L L NL +++ SN ++G+IP + +L LV L+L N LTG+IPS+LG L L L
Sbjct: 29 QLGQLSNLQYLELYSNNMTGKIPDELGNLTNLVSLDLYLNNLTGNIPSTLGNLGKLRFLR 208
Query: 552 LSLNSLSGKV 561
L+ N+LSG +
Sbjct: 209 LNNNTLSGGI 238
>TC9224 similar to GB|AAP21230.1|30102624|BT006422 At3g49750 {Arabidopsis
thaliana;}, partial (77%)
Length = 1318
Score = 61.6 bits (148), Expect = 3e-10
Identities = 34/91 (37%), Positives = 53/91 (57%)
Frame = +1
Query: 471 NDPKLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSN 530
N L++ + SN + LQ+L NL +++S+N++ GEIP + L +++L
Sbjct: 619 NCTNLQALDLSSNFLTGPIPPDLQSLVNLAVLNLSANRLEGEIPPQLTMCAYLNIIDLHQ 798
Query: 531 NLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
NLLTG IP LG L+ L A D+S N L+G +
Sbjct: 799 NLLTGPIPQQLGLLVRLSAFDVSNNRLAGPI 891
Score = 47.4 bits (111), Expect = 6e-06
Identities = 34/111 (30%), Positives = 53/111 (47%), Gaps = 6/111 (5%)
Frame = +1
Query: 457 DPFSQ*VLWQFS------NRNDPKLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKIS 510
DP + L Q S NR + K E+ N S + + N + + +++ +
Sbjct: 415 DPSDEACLTQLSKSLQDPNRLNWKQENFAKPCNDSTSNLQGAICNNGRIYKLSLNNLALH 594
Query: 511 GEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
G I + + L L+LS+N LTG IP L L+NL L+LS N L G++
Sbjct: 595 GTISPFLANCTNLQALDLSSNFLTGPIPPDLQSLVNLAVLNLSANRLEGEI 747
Score = 36.2 bits (82), Expect = 0.014
Identities = 19/40 (47%), Positives = 25/40 (62%)
Frame = +1
Query: 502 IDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSL 541
ID+ N ++G IPQ + L L ++SNN L G IPSSL
Sbjct: 784 IDLHQNLLTGPIPQQLGLLVRLSAFDVSNNRLAGPIPSSL 903
>AV408986
Length = 434
Score = 60.1 bits (144), Expect = 9e-10
Identities = 32/70 (45%), Positives = 44/70 (62%)
Frame = +3
Query: 492 KLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALD 551
+L N N I ID+S N++ G IP+ + + L LL+L N L G IP LG L L+ LD
Sbjct: 168 ELGNCTNAIEIDLSENRLIGIIPKELGQISNLSLLHLFENNLQGHIPRELGSLRQLKKLD 347
Query: 552 LSLNSLSGKV 561
LSLN+L+G +
Sbjct: 348 LSLNNLTGTI 377
Score = 44.3 bits (103), Expect = 5e-05
Identities = 23/53 (43%), Positives = 32/53 (59%)
Frame = +3
Query: 509 ISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
+SGEIP I ++ L LL L N +G+IP LGKL L+ L + N L+G +
Sbjct: 3 LSGEIPPEIGNISSLELLALHQNSFSGAIPKELGKLSGLKRLYVYTNQLNGTI 161
>AV767545
Length = 562
Score = 60.1 bits (144), Expect = 9e-10
Identities = 30/64 (46%), Positives = 46/64 (71%)
Frame = -1
Query: 498 NLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSL 557
+L +++ SN ISG IP I +L L L++S N + G+IPSSLG+L+ L+ LD+S+NSL
Sbjct: 538 SLKVLNLGSNNISGPIPVSISNLIDLERLDISRNHILGAIPSSLGQLLELQWLDVSINSL 359
Query: 558 SGKV 561
+G +
Sbjct: 358 AGSI 347
Score = 53.5 bits (127), Expect = 9e-08
Identities = 31/80 (38%), Positives = 44/80 (54%)
Frame = -1
Query: 482 SNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSL 541
SN + + + NL +L +DIS N I G IP + L L L++S N L GSIPSSL
Sbjct: 514 SNNISGPIPVSISNLIDLERLDISRNHILGAIPSSLGQLLELQWLDVSINSLAGSIPSSL 335
Query: 542 GKLINLEALDLSLNSLSGKV 561
++ NL+ N L G++
Sbjct: 334 SQITNLKHASFRANRLCGEI 275
>BI418931
Length = 516
Score = 58.9 bits (141), Expect = 2e-09
Identities = 27/60 (45%), Positives = 41/60 (68%)
Frame = +2
Query: 502 IDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
+D+S+N + G +P I+ + GL LLNL+ N +G +P+ LGKL+ LE LDLS N +G +
Sbjct: 167 LDVSNNSLEGVLPTEIDKMGGLRLLNLARNGFSGELPNELGKLVYLEYLDLSNNKFTGHI 346
Score = 46.2 bits (108), Expect = 1e-05
Identities = 39/138 (28%), Positives = 65/138 (46%)
Frame = +2
Query: 424 TELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNEYMSN 483
+ L L+LS N+F G ++ G+ S+L ++ P +E + +N
Sbjct: 53 SSLARLNLSGNQFTGPLQLQGSGS---SELLLMSPSQH-------------MEYLDVSNN 184
Query: 484 KGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGK 543
+ ++ + L ++++ N SGE+P + L L L+LSNN TG IP L
Sbjct: 185 SLEGVLPTEIDKMGGLRLLNLARNGFSGELPNELGKLVYLEYLDLSNNKFTGHIPDRLSS 364
Query: 544 LINLEALDLSLNSLSGKV 561
+L A ++S N LSG V
Sbjct: 365 --SLTAFNVSNNDLSGHV 412
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation
aberrant root formation protein), complete
Length = 3308
Score = 57.4 bits (137), Expect = 6e-09
Identities = 32/88 (36%), Positives = 49/88 (55%)
Frame = +2
Query: 471 NDPKLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSN 530
N KL S N + +L ++ +L+++D+S N ++GEIP+ LK L L+N
Sbjct: 905 NLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQ 1084
Query: 531 NLLTGSIPSSLGKLINLEALDLSLNSLS 558
N GS+PS +G L NLE L + N+ S
Sbjct: 1085NKFRGSLPSFIGDLPNLETLQVWENNFS 1168
Score = 55.8 bits (133), Expect = 2e-08
Identities = 25/64 (39%), Positives = 44/64 (68%)
Frame = +2
Query: 498 NLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSL 557
+L A+D+S N ++GE+P+ +++L L +LNLS N ++G +P + + +L LDLS N+
Sbjct: 1703 SLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNF 1882
Query: 558 SGKV 561
+G V
Sbjct: 1883 TGTV 1894
Score = 53.1 bits (126), Expect = 1e-07
Identities = 29/82 (35%), Positives = 53/82 (64%), Gaps = 1/82 (1%)
Frame = +2
Query: 481 MSNKGFS-RVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPS 539
+SN F+ ++ ++NL L ++ + +N+ GEIP + ++ L +N+S N LTG IP+
Sbjct: 1505 LSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPT 1684
Query: 540 SLGKLINLEALDLSLNSLSGKV 561
++ +L A+DLS N+L+G+V
Sbjct: 1685 TITHRASLTAVDLSRNNLAGEV 1750
Score = 51.6 bits (122), Expect = 3e-07
Identities = 49/169 (28%), Positives = 71/169 (41%), Gaps = 31/169 (18%)
Frame = +2
Query: 424 TELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSN---RNDPKLESNEY 480
T LKVL++S+N F G GNI ++L +D + FS KLE +Y
Sbjct: 473 TSLKVLNISHNLFSGQF--PGNITVGMTELEALDAYDN----SFSGPLPEEIVKLEKLKY 634
Query: 481 M---------------------------SNKGFSRVYIKLQNLYNLIAIDIS-SNKISGE 512
+ +N RV L L L + + SN G
Sbjct: 635 LHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGG 814
Query: 513 IPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
IP ++ L LL ++N LTG IP SLG L L +L + +N+L+G +
Sbjct: 815 IPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTI 961
Score = 48.9 bits (115), Expect = 2e-06
Identities = 24/67 (35%), Positives = 45/67 (66%)
Frame = +2
Query: 495 NLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSL 554
++ NL +++++ ++GEIP + +L L L + N LTG+IP L +++L +LDLS+
Sbjct: 833 SMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSI 1012
Query: 555 NSLSGKV 561
N L+G++
Sbjct: 1013NDLTGEI 1033
Score = 37.4 bits (85), Expect = 0.006
Identities = 21/64 (32%), Positives = 35/64 (53%)
Frame = +2
Query: 498 NLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSL 557
+L I +++N + G +P + L + + LSNN L G +PS + +L L LS N
Sbjct: 1346 SLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISG-ESLGTLTLSNNLF 1522
Query: 558 SGKV 561
+GK+
Sbjct: 1523 TGKI 1534
>TC14163
Length = 1712
Score = 56.6 bits (135), Expect = 1e-08
Identities = 47/133 (35%), Positives = 66/133 (49%)
Frame = +1
Query: 429 LSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNEYMSNKGFSR 488
L LS N + SG I + SK+ +D F L N + P GF
Sbjct: 337 LYLSGNPENPKSFFSGTISPSLSKIKNLDGF---YLLNLKNISGPF---------PGFLF 480
Query: 489 VYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLE 548
KLQ +Y I +N++SG IP+ I +L L +L+L+ N TG+IPSS+G L +L
Sbjct: 481 KLPKLQFIY------IENNQLSGRIPENIGNLTRLDVLSLTGNRFTGTIPSSVGGLTHLT 642
Query: 549 ALDLSLNSLSGKV 561
L L NSL+G +
Sbjct: 643 QLQLGNNSLTGTI 681
Score = 52.8 bits (125), Expect = 1e-07
Identities = 47/146 (32%), Positives = 65/146 (44%), Gaps = 5/146 (3%)
Frame = +1
Query: 421 GRFTELKVLSLSNNEFHGDIRCSGNIKCTFSKLHI----IDPFSQ*VLWQFSNRNDPKLE 476
G T L VLSL+ N F G I S ++L + + + + N LE
Sbjct: 550 GNLTRLDVLSLTGNRFTGTIPSSVGGLTHLTQLQLGNNSLTGTIPATIARLKNLTYLSLE 729
Query: 477 SNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKG-LVLLNLSNNLLTG 535
N++ + + +L + +S NK SG+IP I L L L L +N L+G
Sbjct: 730 GNQFSG-----AIPDFFSSFTDLGILRLSRNKFSGKIPASISTLAPKLRYLELGHNQLSG 894
Query: 536 SIPSSLGKLINLEALDLSLNSLSGKV 561
IP LGK L+ LDLS N SG V
Sbjct: 895 KIPDFLGKFRALDTLDLSSNRFSGTV 972
Score = 50.4 bits (119), Expect = 7e-07
Identities = 43/133 (32%), Positives = 60/133 (44%)
Frame = +1
Query: 423 FTELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNEYMS 482
FT+L +L LS N+F SG I + S L PKL E
Sbjct: 772 FTDLGILRLSRNKF------SGKIPASISTLA------------------PKLRYLELGH 879
Query: 483 NKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLG 542
N+ ++ L L +D+SSN+ SG +P ++L + LNL+NNLL P
Sbjct: 880 NQLSGKIPDFLGKFRALDTLDLSSNRFSGTVPASFKNLTKIFNLNLANNLLVDPFPEMNV 1059
Query: 543 KLINLEALDLSLN 555
K +E+LDLS N
Sbjct: 1060K--GIESLDLSNN 1092
>TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1,
partial (27%)
Length = 834
Score = 54.7 bits (130), Expect = 4e-08
Identities = 31/85 (36%), Positives = 47/85 (54%)
Frame = +2
Query: 475 LESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLT 534
L+S + +N +V L NL +++ N++ G IP+ + ++ L +L L N T
Sbjct: 215 LKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWENNFT 394
Query: 535 GSIPSSLGKLINLEALDLSLNSLSG 559
GSIP SLGK L +DLS N L+G
Sbjct: 395 GSIPQSLGKNGKLTLVDLSSNKLTG 469
Score = 50.1 bits (118), Expect = 1e-06
Identities = 29/70 (41%), Positives = 44/70 (62%)
Frame = +2
Query: 492 KLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALD 551
++ NL L+ D + +SGEIP + L+ L L L N+L+GS+ LG L +L+++D
Sbjct: 50 EIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELGHLKSLKSMD 229
Query: 552 LSLNSLSGKV 561
LS N LSG+V
Sbjct: 230 LSNNMLSGQV 259
Score = 42.7 bits (99), Expect = 2e-04
Identities = 22/68 (32%), Positives = 36/68 (52%)
Frame = +2
Query: 493 LQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDL 552
L L L ++ N +SGE P+ + + LSNN L+G +PS++G +++ L L
Sbjct: 629 LFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNFTSMQKLLL 808
Query: 553 SLNSLSGK 560
N SG+
Sbjct: 809 DGNKFSGR 832
Score = 38.9 bits (89), Expect = 0.002
Identities = 38/141 (26%), Positives = 60/141 (41%), Gaps = 2/141 (1%)
Frame = +2
Query: 421 GRFTELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNR--NDPKLESN 478
G L+VL L N F G I S KL ++D S + + +L++
Sbjct: 344 GEMPALEVLQLWENNFTGSIPQSLGKN---GKLTLVDLSSNKLTGTLPPHMCSGNRLQTL 514
Query: 479 EYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIP 538
+ N F + L +L I + N ++G IP+ + L L + +NLL+G P
Sbjct: 515 IALGNFLFGPIPESLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFP 694
Query: 539 SSLGKLINLEALDLSLNSLSG 559
+ N+ + LS N LSG
Sbjct: 695 ETGSVSHNIGQITLSNNKLSG 757
Score = 38.1 bits (87), Expect = 0.004
Identities = 39/164 (23%), Positives = 60/164 (35%), Gaps = 24/164 (14%)
Frame = +2
Query: 421 GRFTELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNEY 480
G LK + LSNN G + S + L++ + +F P LE +
Sbjct: 200 GHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEM-PALEVLQL 376
Query: 481 MSNKGFSRVYIKLQNLYNLIAIDISSNKISGE------------------------IPQV 516
N + L L +D+SSNK++G IP+
Sbjct: 377 WENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPES 556
Query: 517 IEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGK 560
+ + L + + N L GSIP L L L ++ N LSG+
Sbjct: 557 LGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGE 688
Score = 37.7 bits (86), Expect = 0.005
Identities = 22/55 (40%), Positives = 31/55 (56%)
Frame = +2
Query: 507 NKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
N G IP I +L L+ + + L+G IP+ LGKL L+ L L +N LSG +
Sbjct: 23 NNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSL 187
Score = 32.7 bits (73), Expect = 0.16
Identities = 20/63 (31%), Positives = 32/63 (50%)
Frame = +2
Query: 499 LIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLS 558
L + + N +G IPQ + L L++LS+N LTG++P + L+ L N L
Sbjct: 359 LEVLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLF 538
Query: 559 GKV 561
G +
Sbjct: 539 GPI 547
>AV415057
Length = 406
Score = 53.1 bits (126), Expect = 1e-07
Identities = 28/61 (45%), Positives = 39/61 (63%)
Frame = +3
Query: 501 AIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGK 560
A+ + +N++SG IP I L L L+L NN +G+IP + L NLE LDLS N LSG+
Sbjct: 165 ALYLKNNRLSGSIPIEIGQLSVLHQLDLKNNNFSGNIPVQISNLTNLETLDLSGNHLSGE 344
Query: 561 V 561
+
Sbjct: 345 I 347
>BP053628
Length = 567
Score = 52.8 bits (125), Expect = 1e-07
Identities = 28/87 (32%), Positives = 47/87 (53%)
Frame = +1
Query: 475 LESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLT 534
LE+ + SN + + K+ + NL + + N ++G IP + L +L+L NN LT
Sbjct: 67 LEALDLSSNYLYGSIPPKIFTIGNLQTLRLGDNFLNGTIPTLFNSSSNLKVLSLKNNRLT 246
Query: 535 GSIPSSLGKLINLEALDLSLNSLSGKV 561
G PSS+ + L +D+S N +SG +
Sbjct: 247 GQFPSSILSITTLIEIDMSTNEISGSL 327
Score = 41.2 bits (95), Expect = 4e-04
Identities = 38/133 (28%), Positives = 60/133 (44%), Gaps = 1/133 (0%)
Frame = +1
Query: 416 ISILAGRFTELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSN-RNDPK 474
I L + LKVLSL NN G S T ++ + L F+ R+ +
Sbjct: 181 IPTLFNSSSNLKVLSLKNNRLTGQFPSSILSITTLIEIDMSTNEISGSLQDFTGLRSLEQ 360
Query: 475 LESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLT 534
L+ E + ++ +L ++ +S N SG+IP+ LK L L++S N LT
Sbjct: 361 LDLRENRLDSALPKMPPRLMGIF------LSRNSFSGQIPKHYGQLKMLQQLDVSFNALT 522
Query: 535 GSIPSSLGKLINL 547
G+ P+ L L N+
Sbjct: 523 GTXPAELFSLPNI 561
Score = 33.1 bits (74), Expect = 0.12
Identities = 16/27 (59%), Positives = 20/27 (73%)
Frame = +1
Query: 535 GSIPSSLGKLINLEALDLSLNSLSGKV 561
GS+PSS+ +L LEALDLS N L G +
Sbjct: 31 GSLPSSIHRLYALEALDLSSNYLYGSI 111
Score = 27.7 bits (60), Expect = 5.1
Identities = 15/35 (42%), Positives = 22/35 (62%)
Frame = +1
Query: 8 IDLSSSQLYGTLVANSSLFHLVHLQVLDLSDNDFN 42
+DLSS+ LYG++ +F + +LQ L L DN N
Sbjct: 76 LDLSSNYLYGSI--PPKIFTIGNLQTLRLGDNFLN 174
>TC20069 similar to UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3,
partial (14%)
Length = 517
Score = 52.4 bits (124), Expect = 2e-07
Identities = 27/58 (46%), Positives = 36/58 (61%)
Frame = +3
Query: 502 IDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSG 559
+ ++ N +SG IP + + GL LNLSNN G+ PS L L NLE LDL N+L+G
Sbjct: 333 LSLADNGLSGPIPPSLSAVTGLRFLNLSNNGFNGTFPSELSVLKNLEVLDLYNNNLTG 506
Score = 26.9 bits (58), Expect = 8.8
Identities = 14/39 (35%), Positives = 23/39 (58%)
Frame = +3
Query: 523 LVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
++ LNL+ LTG++ + + L L L L+ N LSG +
Sbjct: 252 VIALNLTGLSLTGTLSADVAHLPFLSNLSLADNGLSGPI 368
>TC15223 similar to UP|CAE76632 (CAE76632) Leucine rich repeat protein
precursor, partial (35%)
Length = 876
Score = 52.0 bits (123), Expect = 3e-07
Identities = 27/63 (42%), Positives = 37/63 (57%)
Frame = +3
Query: 499 LIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLS 558
L +++ SN +SG+IP + G+ +LNLS N L G+IP G ALDLS N L
Sbjct: 39 LSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSYFMALDLSYNGLQ 218
Query: 559 GKV 561
G+V
Sbjct: 219 GRV 227
Score = 35.4 bits (80), Expect = 0.025
Identities = 19/51 (37%), Positives = 28/51 (54%)
Frame = +3
Query: 511 GEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
G +P ++ L LNL +N L+G IPS+L + L+LS N L G +
Sbjct: 3 GSVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTI 155
>TC18437 weakly similar to UP|Q9LVB3 (Q9LVB3) Receptor-like protein kinase,
partial (9%)
Length = 503
Score = 51.6 bits (122), Expect = 3e-07
Identities = 47/149 (31%), Positives = 66/149 (43%), Gaps = 3/149 (2%)
Frame = +3
Query: 416 ISILAGRFTELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKL 475
I + G T L L L N G I + CT KL + + + L
Sbjct: 57 IPTVMGNLTRLSELYLHTNRLQGSIPSTLRY-CT--KLQSLGAADNNITGSIPYQTLGYL 227
Query: 476 E---SNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNL 532
E + ++ SN + +L NL +L + + +NK SGEIP + L L L NL
Sbjct: 228 EGLVNLDWSSNSLIGPLPSELGNLKHLSLLYLHANKWSGEIPMELGAG*ALTELMLGRNL 407
Query: 533 LTGSIPSSLGKLINLEALDLSLNSLSGKV 561
GSIPS LG +LE LDLS N+ + +
Sbjct: 408 FRGSIPSFLGSFRSLEILDLSSNNFTSMI 494
Score = 44.7 bits (104), Expect = 4e-05
Identities = 26/66 (39%), Positives = 38/66 (57%)
Frame = +3
Query: 496 LYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLN 555
L NL+ NK+SG IP V+ +L L L L N L GSIPS+L L++L + N
Sbjct: 6 LKNLVRWAWQQNKLSGNIPTVMGNLTRLSELYLHTNRLQGSIPSTLRYCTKLQSLGAADN 185
Query: 556 SLSGKV 561
+++G +
Sbjct: 186 NITGSI 203
Score = 42.4 bits (98), Expect = 2e-04
Identities = 28/81 (34%), Positives = 41/81 (50%), Gaps = 1/81 (1%)
Frame = +3
Query: 480 YMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIP- 538
+ NK + + NL L + + +N++ G IP + L L ++N +TGSIP
Sbjct: 30 WQQNKLSGNIPTVMGNLTRLSELYLHTNRLQGSIPSTLRYCTKLQSLGAADNNITGSIPY 209
Query: 539 SSLGKLINLEALDLSLNSLSG 559
+LG L L LD S NSL G
Sbjct: 210 QTLGYLEGLVNLDWSSNSLIG 272
>AU240163
Length = 300
Score = 51.6 bits (122), Expect = 3e-07
Identities = 27/67 (40%), Positives = 42/67 (62%)
Frame = +1
Query: 493 LQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDL 552
L L +L+ +D+ +N++SG IP+ L+ L LS N ++G IPSS+ ++ L LDL
Sbjct: 97 LTGLRSLMHLDLRNNRLSGPIPRDFGSLRMLSRALLSGNSISGPIPSSISRIYRLADLDL 276
Query: 553 SLNSLSG 559
S N +SG
Sbjct: 277 SRNQVSG 297
Score = 28.9 bits (63), Expect = 2.3
Identities = 15/33 (45%), Positives = 23/33 (69%)
Frame = +1
Query: 529 SNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
++N ++G+IP+SL L +L LDL N LSG +
Sbjct: 61 ADNGISGAIPASLTGLRSLMHLDLRNNRLSGPI 159
>AU089296
Length = 393
Score = 50.8 bits (120), Expect = 6e-07
Identities = 38/133 (28%), Positives = 65/133 (48%)
Frame = +1
Query: 427 KVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNEYMSNKGF 486
++LSLS+N F+G++ + + L +++ +F N KL + N+
Sbjct: 10 RILSLSHNHFYGEVPDLSLL----TNLQVLELDGNAFGPEFPNLGH-KLVALVLRDNRFX 174
Query: 487 SRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLIN 546
S + +L + + L +DIS+N G + L + LN+S N LTG + +L
Sbjct: 175 SGIPAELSSYFQLXRLDISANTFVGPFXTSLLSLPSITYLNISGNKLTGMLFENLSCNSE 354
Query: 547 LEALDLSLNSLSG 559
L+ LDLS N L+G
Sbjct: 355 LQVLDLSSNLLTG 393
>TC14470 similar to UP|AAQ56728 (AAQ56728) Polygalacturonase inhibiting
protein, partial (93%)
Length = 1195
Score = 50.4 bits (119), Expect = 7e-07
Identities = 31/69 (44%), Positives = 40/69 (57%)
Frame = +3
Query: 493 LQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDL 552
+ L NL + ++ ISG IP + +L L L+LS N LTG IPSSL KL NL L L
Sbjct: 342 IAKLTNLRWLILTWTNISGPIPDFLSELPNLQWLHLSFNNLTGPIPSSLSKLPNLIDLRL 521
Query: 553 SLNSLSGKV 561
N L+G +
Sbjct: 522 DRNRLTGPI 548
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.356 0.160 0.547
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,478,251
Number of Sequences: 28460
Number of extensions: 163476
Number of successful extensions: 2181
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 1555
Number of HSP's successfully gapped in prelim test: 67
Number of HSP's that attempted gapping in prelim test: 489
Number of HSP's gapped (non-prelim): 1747
length of query: 561
length of database: 4,897,600
effective HSP length: 95
effective length of query: 466
effective length of database: 2,193,900
effective search space: 1022357400
effective search space used: 1022357400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC137994.10


