

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
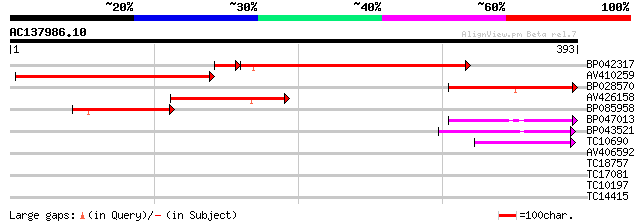
Query= AC137986.10 - phase: 0
(393 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP042317 225 8e-64
AV410259 215 9e-57
BP028570 154 3e-38
AV426158 119 9e-28
BP085958 76 1e-14
BP047013 65 3e-11
BP043521 56 1e-08
TC10690 52 2e-07
AV406592 29 1.2
TC18757 weakly similar to UP|ALAA_ARATH (Q9LI83) Potential phosp... 28 3.4
TC17081 27 4.5
TC10197 similar to UP|Q94AB2 (Q94AB2) AT3g58640/F14P22_230, part... 27 5.9
TC14415 similar to UP|O04237 (O04237) Transcription factor, part... 27 5.9
>BP042317
Length = 551
Score = 225 bits (574), Expect(2) = 8e-64
Identities = 99/165 (60%), Positives = 126/165 (76%), Gaps = 6/165 (3%)
Frame = +3
Query: 161 VTQDGMV------PRDINFIQHTSRLGWKFNKRGKPMIIDPGLYSLNKSDIWWIIKQRNL 214
VTQD ++ RD+NFI+HT GWK + R +P+++DPGLY KSDI W ++R L
Sbjct: 57 VTQDDLLHIFSNLSRDLNFIEHTRIAGWKLSHRARPIVVDPGLYLSKKSDIAWTTQRRTL 236
Query: 215 PTSFKLYTGSAWTIVSRSFSEYCIMGWENLPRTLLLYYTNFVSSPEGYFQTVICNSQEYK 274
PTSFKL+TGSAW +++RS+ EYCI GW NLPRT+L+YYTNF+SS EGYF TVICN++E++
Sbjct: 237 PTSFKLFTGSAWVVLTRSYLEYCIWGWNNLPRTILMYYTNFISSAEGYFHTVICNTEEFR 416
Query: 275 NTTANHDLHYITWDNPPKQHPRSLGLKDYRKMVLSSRPFARKFKR 319
T +HDLHYI WD PPKQHP SL +KD+ KMV S PFARKF +
Sbjct: 417 RTAVSHDLHYIAWDTPPKQHPISLTMKDFDKMVKSKAPFARKFAK 551
Score = 35.4 bits (80), Expect(2) = 8e-64
Identities = 13/18 (72%), Positives = 16/18 (88%)
Frame = +1
Query: 143 KTCHWDWFINLSASDYPL 160
++ W+WFINLSASDYPL
Sbjct: 4 ESSQWNWFINLSASDYPL 57
>AV410259
Length = 418
Score = 215 bits (548), Expect = 9e-57
Identities = 104/139 (74%), Positives = 124/139 (88%), Gaps = 1/139 (0%)
Frame = +1
Query: 5 IFMFTFMVTSILFFFLFIPTRLTLQISTLKPSAMDYFNVLR-TNITYPITFAYLISASKG 63
IFM FM+TS+LFFFLFIPTRLT QIST + +D + +R +N +YP+TFAYLISASKG
Sbjct: 1 IFMVFFMLTSVLFFFLFIPTRLTTQISTFRNVNIDSLSFIRASNSSYPVTFAYLISASKG 180
Query: 64 DTLKLKRLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFSQVGNVWIVGKPNLV 123
D++KLKRLL+VLYHP NYYLIHMDYGAP+AEH+DVAE+VA+DPV+ +VGNVWIVGK NLV
Sbjct: 181 DSVKLKRLLRVLYHPGNYYLIHMDYGAPEAEHRDVAEHVASDPVYGEVGNVWIVGKRNLV 360
Query: 124 TYRGPTMLATTLHAMAMLL 142
TYRGPTMLATTLHA+A+LL
Sbjct: 361 TYRGPTMLATTLHAIAILL 417
>BP028570
Length = 358
Score = 154 bits (388), Expect = 3e-38
Identities = 72/92 (78%), Positives = 82/92 (88%), Gaps = 3/92 (3%)
Frame = -1
Query: 305 KMVLSSRPFARKFKRNNIVLDKIDRDLLKRYKGGFSFGGWCSQGG---RNKACSGLRAEN 361
KMVLS+RPFARKFKRN+ VLD+IDRDLLKRY+G FSFGGWCSQG +N+ACSGLR+EN
Sbjct: 358 KMVLSNRPFARKFKRNDAVLDRIDRDLLKRYRGHFSFGGWCSQGSGGKKNRACSGLRSEN 179
Query: 362 YGLLKPGPGSRRLKNLLNKILMDKFFRQMQCR 393
YGLLKPGP SRRLKNL+ K+L D+FFRQ QCR
Sbjct: 178 YGLLKPGPASRRLKNLITKLLSDRFFRQQQCR 83
>AV426158
Length = 274
Score = 119 bits (298), Expect = 9e-28
Identities = 57/90 (63%), Positives = 71/90 (78%), Gaps = 7/90 (7%)
Frame = +3
Query: 112 GNVWIVGKPNLVTYRGPTMLATTLHAMAMLLKTC-HWDWFINLSASDYPLVTQDGM---- 166
GNV ++ K NLVTYRGPTM+A TLHA A+LL+ C WDWFINLSASDYPLV+QD +
Sbjct: 3 GNVRMIAKANLVTYRGPTMVANTLHAAAILLRECGDWDWFINLSASDYPLVSQDDLLHTF 182
Query: 167 --VPRDINFIQHTSRLGWKFNKRGKPMIID 194
+PRD+NFI HTS +GWK ++R +P+IID
Sbjct: 183 SYLPRDLNFIDHTSDIGWKDHQRARPIIID 272
>BP085958
Length = 507
Score = 75.9 bits (185), Expect = 1e-14
Identities = 38/75 (50%), Positives = 48/75 (63%), Gaps = 4/75 (5%)
Frame = +1
Query: 44 LRTNITYPIT----FAYLISASKGDTLKLKRLLKVLYHPNNYYLIHMDYGAPDAEHKDVA 99
L+ + T PI AYLIS S GD LKR LK LYHP N Y +H+D AP E D+A
Sbjct: 280 LKPSTTSPINSVPRIAYLISGSMGDGETLKRTLKALYHPRNQYAVHLDLEAPPLERLDLA 459
Query: 100 EYVANDPVFSQVGNV 114
+V N+P+F+Q+GNV
Sbjct: 460 NFVKNEPLFAQLGNV 504
>BP047013
Length = 363
Score = 64.7 bits (156), Expect = 3e-11
Identities = 36/89 (40%), Positives = 53/89 (59%)
Frame = -3
Query: 305 KMVLSSRPFARKFKRNNIVLDKIDRDLLKRYKGGFSFGGWCSQGGRNKACSGLRAENYGL 364
KM+ S FARKFK+++ LD I++ L+R G F+ GGWCS G+ K N
Sbjct: 361 KMIASGAAFARKFKQDDPALDLINKKFLRRRNGLFTLGGWCS--GKPKC---TEVGNIYK 197
Query: 365 LKPGPGSRRLKNLLNKILMDKFFRQMQCR 393
LKPGPGS+RL+ L+ ++ + + QC+
Sbjct: 196 LKPGPGSQRLQRLVAELTLKAQSGRDQCK 110
>BP043521
Length = 504
Score = 55.8 bits (133), Expect = 1e-08
Identities = 34/95 (35%), Positives = 49/95 (50%)
Frame = -3
Query: 298 LGLKDYRKMVLSSRPFARKFKRNNIVLDKIDRDLLKRYKGGFSFGGWCSQGGRNKACSGL 357
L D+ M+ S FA+KF+ ++ VLD ID +L R GGWC N C L
Sbjct: 502 LNSTDFDDMIRSGAAFAQKFQPDDPVLDLIDLKILGRSPRSVVPGGWCLGEPGNSTC--L 329
Query: 358 RAENYGLLKPGPGSRRLKNLLNKILMDKFFRQMQC 392
+ +L+PG GS+RL+ + + L + FR QC
Sbjct: 328 TWGDAKILRPGTGSQRLEKAIVEFLSNGTFRSHQC 224
>TC10690
Length = 531
Score = 52.0 bits (123), Expect = 2e-07
Identities = 28/70 (40%), Positives = 38/70 (54%)
Frame = +3
Query: 323 VLDKIDRDLLKRYKGGFSFGGWCSQGGRNKACSGLRAENYGLLKPGPGSRRLKNLLNKIL 382
+LDKID ++L R G+ G W +Q N S N L PGPG+ RLK L+N +L
Sbjct: 9 LLDKIDSEILGRNDHGYVPGKWFTQANPNITKSYSFVRNITELSPGPGAERLKRLINGLL 188
Query: 383 MDKFFRQMQC 392
+ F+ QC
Sbjct: 189 SAENFKNNQC 218
>AV406592
Length = 429
Score = 29.3 bits (64), Expect = 1.2
Identities = 17/55 (30%), Positives = 23/55 (40%)
Frame = +2
Query: 237 CIMGWENLPRTLLLYYTNFVSSPEGYFQTVICNSQEYKNTTANHDLHYITWDNPP 291
C++GW LP TLL Y QT + + Y T + H H I+ P
Sbjct: 275 CVIGWSILPNTLLRTY-----------QTPLLIATYYPTTFSLHSFHTISIQENP 406
>TC18757 weakly similar to UP|ALAA_ARATH (Q9LI83) Potential
phospholipid-transporting ATPase 10 (Aminophospholipid
flippase 10) , partial (15%)
Length = 540
Score = 27.7 bits (60), Expect = 3.4
Identities = 13/42 (30%), Positives = 19/42 (44%), Gaps = 2/42 (4%)
Frame = -1
Query: 253 TNFVSSPEGYFQT--VICNSQEYKNTTANHDLHYITWDNPPK 292
T ++ P+ FQ + C Y N H+L +W N PK
Sbjct: 177 TPLIAQPKIRFQLKRMFCTPS*YNNGNLKHNLEETSWSNTPK 52
>TC17081
Length = 872
Score = 27.3 bits (59), Expect = 4.5
Identities = 11/25 (44%), Positives = 16/25 (64%)
Frame = -2
Query: 156 SDYPLVTQDGMVPRDINFIQHTSRL 180
S +PL+TQDG P+ ++ H S L
Sbjct: 508 SHHPLMTQDGPSPKTLHLFPHYSHL 434
>TC10197 similar to UP|Q94AB2 (Q94AB2) AT3g58640/F14P22_230, partial (13%)
Length = 660
Score = 26.9 bits (58), Expect = 5.9
Identities = 12/27 (44%), Positives = 17/27 (62%)
Frame = -3
Query: 191 MIIDPGLYSLNKSDIWWIIKQRNLPTS 217
++I GL L +S IW + +QRN P S
Sbjct: 247 ILISAGLVGLQESPIWNLREQRNKPPS 167
>TC14415 similar to UP|O04237 (O04237) Transcription factor, partial (83%)
Length = 1666
Score = 26.9 bits (58), Expect = 5.9
Identities = 19/59 (32%), Positives = 31/59 (52%), Gaps = 4/59 (6%)
Frame = -1
Query: 5 IFMFTFMVTSILFFFLFIPTRLTLQISTLKPS----AMDYFNVLRTNITYPITFAYLIS 59
IF+ + ++ LFFF+FI L + IS + S A +F+ +R I++ F L S
Sbjct: 952 IFLQSHLLILRLFFFIFIGRTLLVSISNIFNSRLLIAQIFFSFIRHFISHLSNFITLCS 776
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.325 0.140 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,718,577
Number of Sequences: 28460
Number of extensions: 119009
Number of successful extensions: 933
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 915
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 927
length of query: 393
length of database: 4,897,600
effective HSP length: 92
effective length of query: 301
effective length of database: 2,279,280
effective search space: 686063280
effective search space used: 686063280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC137986.10


