

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137838.7 + phase: 0
(692 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
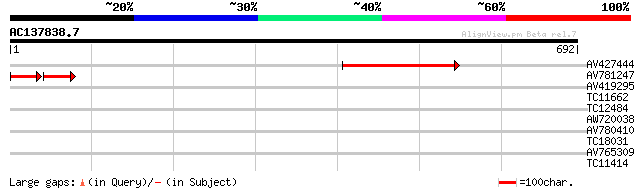
Score E
Sequences producing significant alignments: (bits) Value
AV427444 196 1e-50
AV781247 58 1e-19
AV419295 32 0.34
TC11662 similar to UP|O65748 (O65748) Acyl-CoA synthetase (Frag... 30 1.3
TC12484 29 2.9
AW720038 29 2.9
AV780410 28 4.9
TC18031 similar to UP|Q941L3 (Q941L3) BONZAI1, partial (16%) 28 6.4
AV765309 27 8.3
TC11414 27 8.3
>AV427444
Length = 428
Score = 196 bits (497), Expect = 1e-50
Identities = 109/142 (76%), Positives = 116/142 (80%)
Frame = +2
Query: 407 LEDEFVLWYQVPHLYRLMSWNF*RFALVGE*LKVME*LRLLVL*AALITVTDLVVTLVLQ 466
LEDEFVLW Q P LY L+SWNF*RFALVG *LK E*LR LVL* LI VT+LVV L L
Sbjct: 2 LEDEFVLWCQEPLLYHLISWNF*RFALVGV*LKDTE*LRPLVL*VVLIRVTNLVVMLALL 181
Query: 467 IQPVR*NL*TFRK*TIHLMISPTPAVKFVSGVPLFFKVITKMKLKREKSSMKRDGCIPEI 526
QPVR*NL F+K*TIHLMISP+P VKFVSGVPLF K ITKMKL+REKS MKRDGCI EI
Sbjct: 182 AQPVR*NLWMFQK*TIHLMISPSPVVKFVSGVPLFLKDITKMKLRREKSLMKRDGCIQEI 361
Query: 527 LGHG*LAVVSK*LIGRRISLNW 548
LGHG L VVSK LIG+RIS +W
Sbjct: 362 LGHGYLVVVSKLLIGKRISSSW 427
>AV781247
Length = 297
Score = 58.2 bits (139), Expect(2) = 1e-19
Identities = 28/39 (71%), Positives = 33/39 (83%)
Frame = +2
Query: 1 MDPTPTAQRRLKAINAHLITAVEDNSSDHLQSNPTAGEF 39
MD TP AQRRL+ INAHLI A +D++S HL+SNPTAGEF
Sbjct: 47 MDQTPAAQRRLRTINAHLIAAADDSTS-HLRSNPTAGEF 160
Score = 55.8 bits (133), Expect(2) = 1e-19
Identities = 29/39 (74%), Positives = 31/39 (79%)
Frame = +3
Query: 42 VLYSQRNCRQESGTCIDPYALL*S**TASLIILKLVHCM 80
V YS RNC QESGTCID + L *+**T SLIILK VHCM
Sbjct: 180 VSYSHRNCTQESGTCIDLHVLR*N**TGSLIILK*VHCM 296
>AV419295
Length = 383
Score = 32.0 bits (71), Expect = 0.34
Identities = 14/38 (36%), Positives = 22/38 (57%)
Frame = +2
Query: 221 HHFHHQMEFKLFHIQSFLVRDAAIFSPFAPQSLRMLQP 258
HH HH+ F L ++ ++AA SP +P SL ++ P
Sbjct: 173 HHHHHRQNFPLQLLEKKEDQEAASCSPSSPSSLAIISP 286
>TC11662 similar to UP|O65748 (O65748) Acyl-CoA synthetase (Fragment) ,
partial (81%)
Length = 640
Score = 30.0 bits (66), Expect = 1.3
Identities = 19/59 (32%), Positives = 27/59 (45%)
Frame = +2
Query: 628 LDAKLNLEVLNLQKRSFWWLNHLRWKMVC*LRQ*RLRGLKRKNTLAKQYLICIASSQNP 686
L K + +VL+L ++ HL W + L R RGL NT+ K + C NP
Sbjct: 212 LPRKRS*KVLSL*EQFILTQYHLTWNVTLSLPHTRRRGLSFLNTIRKSLITCTRVEVNP 388
>TC12484
Length = 418
Score = 28.9 bits (63), Expect = 2.9
Identities = 15/47 (31%), Positives = 27/47 (56%), Gaps = 11/47 (23%)
Frame = -2
Query: 144 LTDLSGSLL-----------IMLALHIHTYRCLYMTLSVLMLSNILL 179
L++ SGS++ I++++HIHTY +Y+ S LS+ +L
Sbjct: 393 LSEFSGSMVQPVWSRX**LHILISVHIHTYSFIYINPSYYQLSSSIL 253
>AW720038
Length = 561
Score = 28.9 bits (63), Expect = 2.9
Identities = 11/19 (57%), Positives = 16/19 (83%)
Frame = -3
Query: 250 PQSLRMLQPFAIQVAQLEP 268
P+SL+ML PFAI ++ +EP
Sbjct: 250 PRSLQMLLPFAINISMMEP 194
>AV780410
Length = 548
Score = 28.1 bits (61), Expect = 4.9
Identities = 9/20 (45%), Positives = 14/20 (70%)
Frame = +1
Query: 221 HHFHHQMEFKLFHIQSFLVR 240
HHF HQ +F+ H++ L+R
Sbjct: 244 HHFLHQFDFRFLHLRLLLLR 303
>TC18031 similar to UP|Q941L3 (Q941L3) BONZAI1, partial (16%)
Length = 709
Score = 27.7 bits (60), Expect = 6.4
Identities = 19/41 (46%), Positives = 26/41 (63%), Gaps = 2/41 (4%)
Frame = -3
Query: 197 C*ATCLKFQLCVLLW*LEV*MIKFHHFHH-QME-FKLFHIQ 235
C A LK + VLL LE ++KFHHF H +ME L+H++
Sbjct: 284 CSACKLKIE*VVLLEVLE--LLKFHHFEHPEMEQTGLYHVK 168
>AV765309
Length = 616
Score = 27.3 bits (59), Expect = 8.3
Identities = 10/21 (47%), Positives = 12/21 (56%)
Frame = -2
Query: 221 HHFHHQMEFKLFHIQSFLVRD 241
H FHH + F L H FLV +
Sbjct: 360 HSFHHLLSFPLLHFPCFLVNN 298
>TC11414
Length = 701
Score = 27.3 bits (59), Expect = 8.3
Identities = 15/44 (34%), Positives = 22/44 (49%)
Frame = +1
Query: 640 QKRSFWWLNHLRWKMVC*LRQ*RLRGLKRKNTLAKQYLICIASS 683
Q+RS+W L L +M+C LRG + + Y I +A S
Sbjct: 148 QERSWWMLQELHERMLCQTWTWTLRGWFCRRLIRNYYRIGVADS 279
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.358 0.158 0.552
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,604,566
Number of Sequences: 28460
Number of extensions: 186682
Number of successful extensions: 2670
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 2000
Number of HSP's successfully gapped in prelim test: 64
Number of HSP's that attempted gapping in prelim test: 601
Number of HSP's gapped (non-prelim): 2143
length of query: 692
length of database: 4,897,600
effective HSP length: 97
effective length of query: 595
effective length of database: 2,136,980
effective search space: 1271503100
effective search space used: 1271503100
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC137838.7


