

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137838.4 + phase: 0
(496 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
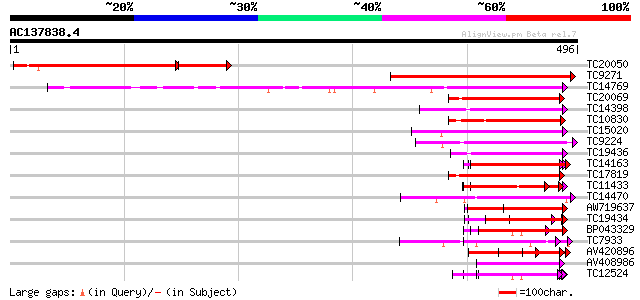
Score E
Sequences producing significant alignments: (bits) Value
TC20050 weakly similar to UP|Q9SFG3 (Q9SFG3) F2O10.5 protein (At... 224 6e-78
TC9271 weakly similar to UP|Q9SFG3 (Q9SFG3) F2O10.5 protein (At3... 277 3e-75
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 155 2e-38
TC20069 similar to UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kina... 86 1e-17
TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%) 76 1e-14
TC10830 weakly similar to UP|Q94JL9 (Q94JL9) At1g68400/T2E12_5, ... 76 1e-14
TC15020 homologue to UP|Q9QX99 (Q9QX99) Transcription factor (T-... 75 2e-14
TC9224 similar to GB|AAP21230.1|30102624|BT006422 At3g49750 {Ara... 74 7e-14
TC19436 similar to UP|Q93Y06 (Q93Y06) Receptor protein kinase-li... 74 7e-14
TC14163 71 4e-13
TC17819 weakly similar to PIR|T05606|T05606 protein kinase homol... 65 3e-11
TC11433 similar to UP|AAR99871 (AAR99871) Strubbelig receptor fa... 64 6e-11
TC14470 similar to UP|AAQ56728 (AAQ56728) Polygalacturonase inhi... 62 2e-10
AW719637 62 2e-10
TC19434 similar to UP|Q94F63 (Q94F63) Somatic embryogenesis rece... 61 5e-10
BP043329 59 2e-09
TC7933 weakly similar to UP|AAQ56728 (AAQ56728) Polygalacturonas... 59 2e-09
AV420896 58 3e-09
AV408986 57 5e-09
TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kina... 57 5e-09
>TC20050 weakly similar to UP|Q9SFG3 (Q9SFG3) F2O10.5 protein (At3g05990),
partial (25%)
Length = 596
Score = 224 bits (570), Expect(2) = 6e-78
Identities = 107/150 (71%), Positives = 126/150 (83%), Gaps = 4/150 (2%)
Frame = +1
Query: 4 LFFLLLFISFHTPSFSQTPP---KGFLINCGTLTTTQINNRTWLPDSNFITTGTPKNITT 60
L FLLLF F S SQTPP +GFLINCG + TQI+ R+WL DS FI+TGTPKN+T+
Sbjct: 4 LIFLLLFSPFSL-SLSQTPPPPPRGFLINCGAPSATQIDGRSWLSDSGFISTGTPKNVTS 180
Query: 61 QVLLPTLKTLRSFPLQVKKHCYN-IPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQIID 119
VL+PTLKTLRSFPL +KKHCYN I V+RG+KY++R+TYFYGGVNG DHP+PPVFDQI+D
Sbjct: 181 PVLIPTLKTLRSFPLNIKKHCYNNIHVFRGSKYLVRSTYFYGGVNGPDHPSPPVFDQIVD 360
Query: 120 GTLWSVVNTTVDYANGNSSFYEGVFLAVGK 149
GTLW+VVNTT DY NGNS+FYEGVFLA G+
Sbjct: 361 GTLWTVVNTTADYVNGNSTFYEGVFLAKGE 450
Score = 84.3 bits (207), Expect(2) = 6e-78
Identities = 38/47 (80%), Positives = 45/47 (94%)
Frame = +2
Query: 148 GKFMSFCIGSNSYTDSDPFVSALEFLILGDSLYNTTDFNNFAIGLVA 194
GKFM+FCIGSN+ T+SDPF+S+LEFLILGDSLYNTTDFNNF++ LVA
Sbjct: 446 GKFMTFCIGSNNRTESDPFISSLEFLILGDSLYNTTDFNNFSMTLVA 586
>TC9271 weakly similar to UP|Q9SFG3 (Q9SFG3) F2O10.5 protein (At3g05990),
partial (32%)
Length = 1035
Score = 277 bits (708), Expect = 3e-75
Identities = 138/163 (84%), Positives = 151/163 (91%), Gaps = 1/163 (0%)
Frame = +1
Query: 334 GPLINAGEVFNVLSLGGRTSTRDVIALQRVKESLRNPPLDWSGDPCVPRQYSWTGITCSE 393
G INAGE+F+V SLGG TS RDVIAL +VKESLRNPPLDWSGDPC+PRQYSWTGITCSE
Sbjct: 7 GTRINAGEIFDVRSLGGTTSIRDVIALVKVKESLRNPPLDWSGDPCMPRQYSWTGITCSE 186
Query: 394 GLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPNLSSLTMLETLHLEE 453
G RIR+VTLNLT+M+LSGSLS FVANMTAL NIWLGNNSLSGQIP LSSLT+LETLHLE+
Sbjct: 187 GPRIRVVTLNLTNMNLSGSLSPFVANMTALANIWLGNNSLSGQIPELSSLTLLETLHLED 366
Query: 454 NQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANLL-KPGLSI 495
NQF+GEIPSSLGNIS+L EVFLQNNNLTGQIPANL+ KPGLS+
Sbjct: 367 NQFTGEIPSSLGNISTLHEVFLQNNNLTGQIPANLIGKPGLSL 495
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 155 bits (391), Expect = 2e-38
Identities = 133/472 (28%), Positives = 213/472 (44%), Gaps = 17/472 (3%)
Frame = +3
Query: 34 TTTQINNRTWLPDSNFITTGTPKNITTQVLLPTLKTLRSFPLQVKKHCYNIPVYRGAKYM 93
T + TW D K T++ + + +R F + K CY++P + Y+
Sbjct: 531 TLNYTTDYTWFSDKR----SCRKIPETELRNRSNENVRLFDIDEGKRCYDLPTIKNGVYL 698
Query: 94 IRTTYFYGGVNGVDHPTPPVFDQIIDGTLWSVVNTTVDYANGNSSFYEGVFLAVGKFMSF 153
IR T+ + +N F+ I T V ++ EGVF A ++ F
Sbjct: 699 IRGTFPFDSLNSS-------FNASIGVTQLGAVRSS----RLQDLEIEGVFRATKDYIDF 845
Query: 154 CIGSNSYTDSDPFVSALEFLILGDSLYNTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFD 213
C+ PF+S LE + DF + L++RN+ G + IR+P DQ D
Sbjct: 846 CLLKGEVY---PFISQLELRPSPEEYLQ--DFPTSVLKLISRNNLGDTKDDIRFPVDQSD 1010
Query: 214 RIWEPFGQSNST---KANTENVSVSGFWNLPPSKVFETHLGSEQLESLELRWPTASLPSS 270
RIW+ S+S +N NV ++ PP V +T L E LE
Sbjct: 1011RIWKASSISSSAVPLSSNVSNVDLNANVT-PPLTVLQTALTDP--ERLEFIHTDLETEDY 1181
Query: 271 KYYIALYF--ADNT--AGSRIFNISVNGVHYYRDLNAIASGVVVFANQWPL--SGPTTIT 324
Y + LYF D T AG R+F+I VN + +A G + + SG +T
Sbjct: 1182GYRVFLYFLELDRTLQAGQRVFDIYVNSEIKKESFDVLAGGSNYRYDVLDISASGSLNVT 1361
Query: 325 LTPSASSSLGPLINAGEVFNVLSLGGRTSTRDVIALQRVKESL-----RNPPLD-WSGDP 378
L ++ S GPL+NA E+ V T+ DV +Q+++E L N L+ WSGDP
Sbjct: 1362LVKASKSEFGPLLNAYEILQVRPWIEETNQTDVGVIQKMREELLLQNSGNRALESWSGDP 1541
Query: 379 CVPRQYSWTGITCSEGLRIRIVT-LNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQI 437
C+ W GI C ++T L+L+S +L G + S +A MT L + + +NS G +
Sbjct: 1542CI--LLPWKGIACDGSNGSSVITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSV 1715
Query: 438 PNLSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQ-NNNLTGQIPANL 488
P+ ++L ++ L N G++P S+ + LK ++ N +++ + PAN+
Sbjct: 1716PSFPLSSLLISVDLSYNDLMGKLPESIVKLPHLKSLYFGCNEHMSPEDPANM 1871
>TC20069 similar to UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3,
partial (14%)
Length = 517
Score = 86.3 bits (212), Expect = 1e-17
Identities = 46/102 (45%), Positives = 69/102 (67%), Gaps = 1/102 (0%)
Frame = +3
Query: 385 SWTGITCSEGLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIP-NLSSL 443
SW+G+TC R ++ LNLT + L+G+LS+ VA++ L+N+ L +N LSG IP +LS++
Sbjct: 216 SWSGVTCDP--RRHVIALNLTGLSLTGTLSADVAHLPFLSNLSLADNGLSGPIPPSLSAV 389
Query: 444 TMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIP 485
T L L+L N F+G PS L + +L+ + L NNNLTG +P
Sbjct: 390 TGLRFLNLSNNGFNGTFPSELSVLKNLEVLDLYNNNLTGVLP 515
>TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%)
Length = 982
Score = 75.9 bits (185), Expect = 1e-14
Identities = 50/132 (37%), Positives = 76/132 (56%), Gaps = 2/132 (1%)
Frame = +1
Query: 359 ALQRVKESLRNPP-LDWSGDPCVPRQYSWTGITCSEGLRIRIVTLNLTSMDLSGSLSSFV 417
AL K+SL +P + S D + +W +TC + R+ +L +++LSG L +
Sbjct: 214 ALYAFKQSLSDPDNVLQSWDATLVSPCTWFHVTCQDNSVTRV---DLGNLNLSGHLVPDL 384
Query: 418 ANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQ 476
N+ +L + L N++ G IP L +L L +L L N SG IPSSLGN+ +L+ + L
Sbjct: 385 GNLHSLQYLELYENNIQGTIPEELGNLQSLISLDLYHNNVSGSIPSSLGNLKNLRFLRLN 564
Query: 477 NNNLTGQIPANL 488
NN+LTGQIP +L
Sbjct: 565 NNHLTGQIPKSL 600
>TC10830 weakly similar to UP|Q94JL9 (Q94JL9) At1g68400/T2E12_5, partial
(24%)
Length = 702
Score = 75.9 bits (185), Expect = 1e-14
Identities = 43/102 (42%), Positives = 64/102 (62%)
Frame = +2
Query: 385 SWTGITCSEGLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPNLSSLT 444
+W G+TC LR R+ L L ++DL GSL + ++T L + L NN G IPNLS+LT
Sbjct: 290 TWHGVTC---LRNRVSRLVLENLDLHGSLHP-LTSLTQLRVLSLKNNRFHGPIPNLSNLT 457
Query: 445 MLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPA 486
+ L L N FSG+ P +L +++ L + L +N+L+G+IPA
Sbjct: 458 GMRLLFLSHNNFSGDFPVTLTSLTRLYRLDLSHNSLSGEIPA 583
>TC15020 homologue to UP|Q9QX99 (Q9QX99) Transcription factor (T-cell
leukemia, homeobox 1), partial (5%)
Length = 1304
Score = 75.1 bits (183), Expect = 2e-14
Identities = 52/150 (34%), Positives = 81/150 (53%), Gaps = 13/150 (8%)
Frame = +1
Query: 352 TSTRDVIALQRVKESLRNPPL-DWSG--------DPC-VPRQYSWT-GITCSEGLRIRIV 400
TS D+ AL+ K S++ + WS DPC +PR+ + G+TC+ +
Sbjct: 58 TSASDIAALKAFKASIKPSSIAPWSCLASWNFTIDPCSLPRRTHFICGLTCTATTTTNNI 237
Query: 401 T-LNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSG 458
+ L SGSL+ ++ +T L + L NS SG IP+ LSSL+ L+TL L N FSG
Sbjct: 238 NQITLDPAGYSGSLTPLISQLTQLVTLDLAENSFSGPIPSSLSSLSNLKTLTLRSNSFSG 417
Query: 459 EIPSSLGNISSLKEVFLQNNNLTGQIPANL 488
IP S+ + SL + +N+L+G +P ++
Sbjct: 418 SIPPSITTLKSLYSLDFSHNSLSGSLPNSM 507
>TC9224 similar to GB|AAP21230.1|30102624|BT006422 At3g49750 {Arabidopsis
thaliana;}, partial (77%)
Length = 1318
Score = 73.6 bits (179), Expect = 7e-14
Identities = 54/147 (36%), Positives = 78/147 (52%), Gaps = 6/147 (4%)
Frame = +1
Query: 356 DVIALQRVKESLRNPP-LDWSGD----PCVPRQYSWTGITCSEGLRIRIVTLNLTSMDLS 410
D L ++ +SL++P L+W + PC + G C+ G RI L+L ++ L
Sbjct: 424 DEACLTQLSKSLQDPNRLNWKQENFAKPCNDSTSNLQGAICNNG---RIYKLSLNNLALH 594
Query: 411 GSLSSFVANMTALTNIWLGNNSLSGQIP-NLSSLTMLETLHLEENQFSGEIPSSLGNISS 469
G++S F+AN T L + L +N L+G IP +L SL L L+L N+ GEIP L +
Sbjct: 595 GTISPFLANCTNLQALDLSSNFLTGPIPPDLQSLVNLAVLNLSANRLEGEIPPQLTMCAY 774
Query: 470 LKEVFLQNNNLTGQIPANLLKPGLSIR 496
L + L N LTG IP L GL +R
Sbjct: 775 LNIIDLHQNLLTGPIPQQL---GLLVR 846
>TC19436 similar to UP|Q93Y06 (Q93Y06) Receptor protein kinase-like protein,
partial (19%)
Length = 570
Score = 73.6 bits (179), Expect = 7e-14
Identities = 41/104 (39%), Positives = 62/104 (59%), Gaps = 1/104 (0%)
Frame = +1
Query: 386 WTGITCSEGLRIRIVTLNLTSMDLSGSL-SSFVANMTALTNIWLGNNSLSGQIPNLSSLT 444
W G+ CS+G R V L + L+G+ + + + L + L NNSL+G IP+LS LT
Sbjct: 220 WQGVKCSQGRVFRFV---LQDLSLAGTFPADTLTRLDQLRVLTLRNNSLTGPIPDLSPLT 390
Query: 445 MLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANL 488
L++L L+ N FSG P S+ ++ L + L +NNL+GQ+P L
Sbjct: 391 NLKSLFLDRNHFSGAFPPSILSLHRLLTLSLSHNNLSGQLPVQL 522
>TC14163
Length = 1712
Score = 71.2 bits (173), Expect = 4e-13
Identities = 37/88 (42%), Positives = 57/88 (64%), Gaps = 1/88 (1%)
Frame = +1
Query: 404 LTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIP-NLSSLTMLETLHLEENQFSGEIPS 462
L ++SG F+ + L I++ NN LSG+IP N+ +LT L+ L L N+F+G IPS
Sbjct: 436 LNLKNISGPFPGFLFKLPKLQFIYIENNQLSGRIPENIGNLTRLDVLSLTGNRFTGTIPS 615
Query: 463 SLGNISSLKEVFLQNNNLTGQIPANLLK 490
S+G ++ L ++ L NN+LTG IPA + +
Sbjct: 616 SVGGLTHLTQLQLGNNSLTGTIPATIAR 699
Score = 67.0 bits (162), Expect = 7e-12
Identities = 38/92 (41%), Positives = 55/92 (59%), Gaps = 1/92 (1%)
Frame = +1
Query: 398 RIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIP-NLSSLTMLETLHLEENQF 456
R+ L+LT +G++ S V +T LT + LGNNSL+G IP ++ L L L LE NQF
Sbjct: 562 RLDVLSLTGNRFTGTIPSSVGGLTHLTQLQLGNNSLTGTIPATIARLKNLTYLSLEGNQF 741
Query: 457 SGEIPSSLGNISSLKEVFLQNNNLTGQIPANL 488
SG IP + + L + L N +G+IPA++
Sbjct: 742 SGAIPDFFSSFTDLGILRLSRNKFSGKIPASI 837
Score = 51.6 bits (122), Expect = 3e-07
Identities = 30/86 (34%), Positives = 48/86 (54%), Gaps = 2/86 (2%)
Frame = +1
Query: 402 LNLTSMDLSGSLSSFVANMTA-LTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSGE 459
L L+ SG + + ++ + L + LG+N LSG+IP+ L L+TL L N+FSG
Sbjct: 790 LRLSRNKFSGKIPASISTLAPKLRYLELGHNQLSGKIPDFLGKFRALDTLDLSSNRFSGT 969
Query: 460 IPSSLGNISSLKEVFLQNNNLTGQIP 485
+P+S N++ + + L NN L P
Sbjct: 970 VPASFKNLTKIFNLNLANNLLVDPFP 1047
>TC17819 weakly similar to PIR|T05606|T05606 protein kinase homolog
F9D16.210 - Arabidopsis thaliana {Arabidopsis
thaliana;}, partial (9%)
Length = 538
Score = 65.1 bits (157), Expect = 3e-11
Identities = 35/103 (33%), Positives = 64/103 (61%), Gaps = 2/103 (1%)
Frame = +1
Query: 385 SWTGITCSEGLRIRIVTLNLTSMDLSGSLS-SFVANMTALTNIWLGNNSLSGQIP-NLSS 442
+W G+ C + R + +L L ++ L G+L + ++ ++ L + +N L+G+IP + S+
Sbjct: 226 NWVGVQC-DASRSFVYSLRLPAVGLVGNLPPNTISRLSQLRVLSFRSNGLTGEIPADFSN 402
Query: 443 LTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIP 485
LT L +L+L++NQ SGE P SL ++ L + L +NN +G +P
Sbjct: 403 LTFLRSLYLQKNQLSGEFPPSLTRLTRLNRLDLSSNNFSGAVP 531
Score = 28.1 bits (61), Expect = 3.4
Identities = 10/27 (37%), Positives = 20/27 (74%)
Frame = +1
Query: 461 PSSLGNISSLKEVFLQNNNLTGQIPAN 487
P+++ +S L+ + ++N LTG+IPA+
Sbjct: 313 PNTISRLSQLRVLSFRSNGLTGEIPAD 393
>TC11433 similar to UP|AAR99871 (AAR99871) Strubbelig receptor family 3,
partial (24%)
Length = 565
Score = 63.9 bits (154), Expect = 6e-11
Identities = 34/82 (41%), Positives = 52/82 (62%), Gaps = 1/82 (1%)
Frame = +2
Query: 404 LTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSGEIPS 462
L + +GS+ + ++ +T LT++ L N LSG+IP+ SLT L L L N SGE+P
Sbjct: 200 LAANQFTGSIPTSLSTLTGLTDMSLNENHLSGEIPDAFQSLTQLINLDLSTNNLSGELPP 379
Query: 463 SLGNISSLKEVFLQNNNLTGQI 484
S+ N+S+L + LQNN L+G +
Sbjct: 380 SVENLSALTTLRLQNNQLSGTL 445
Score = 57.4 bits (137), Expect = 5e-09
Identities = 31/93 (33%), Positives = 56/93 (59%), Gaps = 1/93 (1%)
Frame = +2
Query: 397 IRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIP-NLSSLTMLETLHLEENQ 455
+ I ++L++ ++ GS+ S + + N +L N +G IP +LS+LT L + L EN
Sbjct: 113 VTISVIDLSNNNIGGSIPSSLP--VTMRNFFLAANQFTGSIPTSLSTLTGLTDMSLNENH 286
Query: 456 FSGEIPSSLGNISSLKEVFLQNNNLTGQIPANL 488
SGEIP + +++ L + L NNL+G++P ++
Sbjct: 287 LSGEIPDAFQSLTQLINLDLSTNNLSGELPPSV 385
Score = 57.0 bits (136), Expect = 7e-09
Identities = 31/75 (41%), Positives = 48/75 (63%)
Frame = +2
Query: 398 RIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPNLSSLTMLETLHLEENQFS 457
+++ L+L++ +LSG L V N++ALT + L NN LSG + L L L+ L++E NQF+
Sbjct: 326 QLINLDLSTNNLSGELPPSVENLSALTTLRLQNNQLSGTLDVLQDLP-LQDLNVENNQFA 502
Query: 458 GEIPSSLGNISSLKE 472
G IP L NI ++
Sbjct: 503 GPIPPKLLNIPKFRQ 547
Score = 39.7 bits (91), Expect = 0.001
Identities = 24/71 (33%), Positives = 36/71 (49%), Gaps = 1/71 (1%)
Frame = +2
Query: 419 NMTALTNIWLGNNSLSGQI-PNLSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQN 477
N + + I L +L G++ NLS+ + + L N G IPSSL +++ FL
Sbjct: 35 NGSFIQKIVLNGANLGGELGDNLSTFVTISVIDLSNNNIGGSIPSSLP--VTMRNFFLAA 208
Query: 478 NNLTGQIPANL 488
N TG IP +L
Sbjct: 209 NQFTGSIPTSL 241
>TC14470 similar to UP|AAQ56728 (AAQ56728) Polygalacturonase inhibiting
protein, partial (93%)
Length = 1195
Score = 62.4 bits (150), Expect = 2e-10
Identities = 51/185 (27%), Positives = 84/185 (44%), Gaps = 32/185 (17%)
Frame = +3
Query: 343 FNVLSLGGRTSTRDVIALQRVKESLRNPPL-----------DWSGDPCVPRQYSWTGITC 391
F+ ++ GR + D L ++K+ L NP L DW C P+ + T +
Sbjct: 39 FSHVAFSGRCNPEDKKVLLQIKKDLNNPYLLASWNPETACCDWYCVECDPKTHRITSLII 218
Query: 392 SEGLR-----------------IRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLS 434
S + ++++ + L+G + +A +T L + L ++S
Sbjct: 219 SSSVPETNFSGQIPPSVGDLPYLQVLEFHKLPK-LTGPIQPAIAKLTNLRWLILTWTNIS 395
Query: 435 GQIPN-LSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIP---ANLLK 490
G IP+ LS L L+ LHL N +G IPSSL + +L ++ L N LTG IP + K
Sbjct: 396 GPIPDFLSELPNLQWLHLSFNNLTGPIPSSLSKLPNLIDLRLDRNRLTGPIPDSFGSFKK 575
Query: 491 PGLSI 495
PG+ +
Sbjct: 576 PGIDL 590
>AW719637
Length = 571
Score = 62.0 bits (149), Expect = 2e-10
Identities = 33/91 (36%), Positives = 50/91 (54%), Gaps = 1/91 (1%)
Frame = +2
Query: 399 IVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFS 457
+V L L +SGSL S + + + I + LSG IP + + + L+ L+L +N S
Sbjct: 263 LVMLGLAETSISGSLPSSIQLLKRIKTIAIYTTLLSGSIPEEIGNCSELQNLYLYQNSIS 442
Query: 458 GEIPSSLGNISSLKEVFLQNNNLTGQIPANL 488
G IPS +G +S LK + L NN+ G IP +
Sbjct: 443 GSIPSQIGELSKLKSLLLWQNNIVGTIPEEI 535
Score = 53.5 bits (127), Expect = 8e-08
Identities = 30/90 (33%), Positives = 56/90 (61%), Gaps = 2/90 (2%)
Frame = +2
Query: 401 TLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIP-NLSSLTMLETLHLEENQ-FSG 458
+L+L + L G++ S + N+++L N+ L +N LSG+IP ++ SL L+ N+ G
Sbjct: 50 SLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSIGSLRNLQVFRAGGNKNLKG 229
Query: 459 EIPSSLGNISSLKEVFLQNNNLTGQIPANL 488
EIP +GN +SL + L +++G +P+++
Sbjct: 230 EIPWEIGNCTSLVMLGLAETSISGSLPSSI 319
Score = 48.9 bits (115), Expect = 2e-06
Identities = 25/57 (43%), Positives = 40/57 (69%), Gaps = 1/57 (1%)
Frame = +2
Query: 433 LSGQIPN-LSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANL 488
L G+IP + SL+ L++L L N G IPS++GN+SSL + L +N+L+G+IP ++
Sbjct: 2 LLGEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSI 172
>TC19434 similar to UP|Q94F63 (Q94F63) Somatic embryogenesis receptor-like
kinase 2, partial (26%)
Length = 550
Score = 60.8 bits (146), Expect = 5e-10
Identities = 35/87 (40%), Positives = 52/87 (59%), Gaps = 1/87 (1%)
Frame = +2
Query: 402 LNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSGEI 460
L L S +++G + + N+T L ++ L N+L+G IP+ L +L L L L N SG I
Sbjct: 59 LELYSNNMTGKIPDELGNLTNLVSLDLYLNNLTGNIPSTLGNLGKLRFLRLNNNTLSGGI 238
Query: 461 PSSLGNISSLKEVFLQNNNLTGQIPAN 487
P +L N+SSL+ + L NN L G +P N
Sbjct: 239 PMTLTNVSSLQVLDLSNNQLKGVVPVN 319
Score = 52.8 bits (125), Expect = 1e-07
Identities = 29/73 (39%), Positives = 47/73 (63%), Gaps = 1/73 (1%)
Frame = +2
Query: 417 VANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFL 475
+ ++ L + L +N+++G+IP+ L +LT L +L L N +G IPS+LGN+ L+ + L
Sbjct: 32 LGQLSNLQYLELYSNNMTGKIPDELGNLTNLVSLDLYLNNLTGNIPSTLGNLGKLRFLRL 211
Query: 476 QNNNLTGQIPANL 488
NN L+G IP L
Sbjct: 212 NNNTLSGGIPMTL 250
Score = 48.9 bits (115), Expect = 2e-06
Identities = 29/81 (35%), Positives = 49/81 (59%), Gaps = 1/81 (1%)
Frame = +2
Query: 399 IVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIP-NLSSLTMLETLHLEENQFS 457
+V+L+L +L+G++ S + N+ L + L NN+LSG IP L++++ L+ L L NQ
Sbjct: 122 LVSLDLYLNNLTGNIPSTLGNLGKLRFLRLNNNTLSGGIPMTLTNVSSLQVLDLSNNQLK 301
Query: 458 GEIPSSLGNISSLKEVFLQNN 478
G +P + G+ S + QNN
Sbjct: 302 GVVPVN-GSFSLFTPISYQNN 361
Score = 47.8 bits (112), Expect = 4e-06
Identities = 23/51 (45%), Positives = 32/51 (62%)
Frame = +2
Query: 438 PNLSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANL 488
P L L+ L+ L L N +G+IP LGN+++L + L NNLTG IP+ L
Sbjct: 26 PQLGQLSNLQYLELYSNNMTGKIPDELGNLTNLVSLDLYLNNLTGNIPSTL 178
>BP043329
Length = 484
Score = 58.9 bits (141), Expect = 2e-09
Identities = 32/86 (37%), Positives = 49/86 (56%), Gaps = 1/86 (1%)
Frame = +3
Query: 404 LTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSGEIPS 462
LT +L G + + N+ L ++ L N L G IP+ L+ LT L + L N SGE+P
Sbjct: 126 LTQCNLVGVIPDSIGNLKKLKDLDLALNDLYGSIPSSLTGLTSLRQIELYNNSLSGELPR 305
Query: 463 SLGNISSLKEVFLQNNNLTGQIPANL 488
+GN++ L+ + N+LTG+IP L
Sbjct: 306 GMGNLTELRLLDASMNHLTGRIPEEL 383
Score = 55.8 bits (133), Expect = 2e-08
Identities = 29/79 (36%), Positives = 48/79 (60%), Gaps = 1/79 (1%)
Frame = +3
Query: 411 GSLSSFVANMTALTNIWLGNNSLSGQIP-NLSSLTMLETLHLEENQFSGEIPSSLGNISS 469
G + + N+T L +WL +L G IP ++ +L L+ L L N G IPSSL ++S
Sbjct: 75 GRIPPEIGNLTNLEVVWLTQCNLVGVIPDSIGNLKKLKDLDLALNDLYGSIPSSLTGLTS 254
Query: 470 LKEVFLQNNNLTGQIPANL 488
L+++ L NN+L+G++P +
Sbjct: 255 LRQIELYNNSLSGELPRGM 311
Score = 51.2 bits (121), Expect = 4e-07
Identities = 34/100 (34%), Positives = 54/100 (54%), Gaps = 24/100 (24%)
Frame = +3
Query: 398 RIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIP----NLSSLTML------- 446
++ L+L DL GS+ S + +T+L I L NNSLSG++P NL+ L +L
Sbjct: 180 KLKDLDLALNDLYGSIPSSLTGLTSLRQIELYNNSLSGELPRGMGNLTELRLLDASMNHL 359
Query: 447 -------------ETLHLEENQFSGEIPSSLGNISSLKEV 473
E+L+L EN+F GE+P+S+ + +L E+
Sbjct: 360 TGRIPEELCSLPLESLNLYENRFEGELPASIADSPNLYEL 479
>TC7933 weakly similar to UP|AAQ56728 (AAQ56728) Polygalacturonase
inhibiting protein, partial (78%)
Length = 1320
Score = 58.5 bits (140), Expect = 2e-09
Identities = 51/186 (27%), Positives = 83/186 (44%), Gaps = 35/186 (18%)
Frame = +3
Query: 342 VFNVLSLGGRTSTRDVIALQRVKESLRNPPLDWSGDP-----------CVPRQYSWTGIT 390
+F+ ++ GR +D L ++K+ +P L S DP C P+ + T +
Sbjct: 48 LFSPVAFSGRCHPQDKKVLLQIKKDFNSPYLLASWDPKTACCDWYCVECDPKTHRITSLI 227
Query: 391 CSEGLRIRIVTLNLTSM--------------------DLSGSLSSFVANMTALTNIWLGN 430
S I NL+ L+G + +A +T L ++ +
Sbjct: 228 ISSS----IPDTNLSGPIPPSVGDLPYLEYLDFHKLPKLTGPIQPAIAKLTKLKSLSITW 395
Query: 431 NSLSGQIPN-LSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPAN-- 487
++SG IP+ L+ L L++L+L N SG IP+SL + +L + L N LTG IPA+
Sbjct: 396 TNVSGPIPDFLAQLKNLDSLYLSFNNLSGHIPASLSRLPNLLSLNLDRNKLTGPIPASFG 575
Query: 488 -LLKPG 492
KPG
Sbjct: 576 FFKKPG 593
Score = 46.2 bits (108), Expect = 1e-05
Identities = 34/111 (30%), Positives = 55/111 (48%), Gaps = 26/111 (23%)
Frame = +3
Query: 398 RIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIP-NLSSLTMLETLHLEEN-- 454
++ +L++T ++SG + F+A + L +++L N+LSG IP +LS L L +L+L+ N
Sbjct: 369 KLKSLSITWTNVSGPIPDFLAQLKNLDSLYLSFNNLSGHIPASLSRLPNLLSLNLDRNKL 548
Query: 455 -----------------------QFSGEIPSSLGNISSLKEVFLQNNNLTG 482
Q SG IPSSLG + + + N LTG
Sbjct: 549 TGPIPASFGFFKKPGPNLFLSHNQLSGPIPSSLGQLDP-ERIDFSRNKLTG 698
>AV420896
Length = 203
Score = 58.2 bits (139), Expect = 3e-09
Identities = 27/63 (42%), Positives = 44/63 (68%)
Frame = +3
Query: 402 LNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPNLSSLTMLETLHLEENQFSGEIP 461
+NL++ +GS+ ++N+T L ++ L NNSLSG+IP+L+ T L+ L+LE N +G +P
Sbjct: 6 INLSNNSFNGSIPISISNLTHLNSLVLANNSLSGEIPDLNVPTTLQELNLENNNLTGVVP 185
Query: 462 SSL 464
SL
Sbjct: 186 KSL 194
Score = 56.6 bits (135), Expect = 9e-09
Identities = 32/64 (50%), Positives = 45/64 (70%), Gaps = 1/64 (1%)
Frame = +3
Query: 428 LGNNSLSGQIP-NLSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPA 486
L NNS +G IP ++S+LT L +L L N SGEIP L ++L+E+ L+NNNLTG +P
Sbjct: 12 LSNNSFNGSIPISISNLTHLNSLVLANNSLSGEIPD-LNVPTTLQELNLENNNLTGVVPK 188
Query: 487 NLLK 490
+LL+
Sbjct: 189 SLLR 200
Score = 40.4 bits (93), Expect = 7e-04
Identities = 16/37 (43%), Positives = 26/37 (70%)
Frame = +3
Query: 449 LHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIP 485
++L N F+G IP S+ N++ L + L NN+L+G+IP
Sbjct: 6 INLSNNSFNGSIPISISNLTHLNSLVLANNSLSGEIP 116
>AV408986
Length = 434
Score = 57.4 bits (137), Expect = 5e-09
Identities = 33/78 (42%), Positives = 44/78 (56%), Gaps = 1/78 (1%)
Frame = +3
Query: 409 LSGSLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSGEIPSSLGNI 467
L+G++ + + N T I L N L G IP L ++ L LHL EN G IP LG++
Sbjct: 147 LNGTIPTELGNCTNAIEIDLSENRLIGIIPKELGQISNLSLLHLFENNLQGHIPRELGSL 326
Query: 468 SSLKEVFLQNNNLTGQIP 485
LK++ L NNLTG IP
Sbjct: 327 RQLKKLDLSLNNLTGTIP 380
Score = 37.0 bits (84), Expect = 0.007
Identities = 18/34 (52%), Positives = 23/34 (66%)
Frame = +3
Query: 457 SGEIPSSLGNISSLKEVFLQNNNLTGQIPANLLK 490
SGEIP +GNISSL+ + L N+ +G IP L K
Sbjct: 6 SGEIPPEIGNISSLELLALHQNSFSGAIPKELGK 107
>TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1,
partial (27%)
Length = 834
Score = 57.4 bits (137), Expect = 5e-09
Identities = 41/116 (35%), Positives = 58/116 (49%), Gaps = 25/116 (21%)
Frame = +2
Query: 398 RIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIP----NLSSLTML------- 446
++ TL L LSGSL+ + ++ +L ++ L NN LSGQ+P L +LT+L
Sbjct: 140 KLDTLFLQVNVLSGSLTPELGHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRL 319
Query: 447 --------------ETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANL 488
E L L EN F+G IP SLG L V L +N LTG +P ++
Sbjct: 320 HGAIPEFVGEMPALEVLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHM 487
Score = 55.8 bits (133), Expect = 2e-08
Identities = 31/78 (39%), Positives = 44/78 (55%), Gaps = 1/78 (1%)
Frame = +2
Query: 411 GSLSSFVANMTALTNIWLGNNSLSGQIP-NLSSLTMLETLHLEENQFSGEIPSSLGNISS 469
G + + N+T L LSG+IP L L L+TL L+ N SG + LG++ S
Sbjct: 35 GGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELGHLKS 214
Query: 470 LKEVFLQNNNLTGQIPAN 487
LK + L NN L+GQ+PA+
Sbjct: 215 LKSMDLSNNMLSGQVPAS 268
Score = 48.1 bits (113), Expect = 3e-06
Identities = 23/76 (30%), Positives = 45/76 (58%), Gaps = 1/76 (1%)
Frame = +2
Query: 409 LSGSLSSFVANMTALTNIWLGNNSLSGQIPNLSSLTM-LETLHLEENQFSGEIPSSLGNI 467
L+GS+ + + LT + +N LSG+ P S++ + + L N+ SG +PS++GN
Sbjct: 605 LNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNF 784
Query: 468 SSLKEVFLQNNNLTGQ 483
+S++++ L N +G+
Sbjct: 785 TSMQKLLLDGNKFSGR 832
Score = 47.4 bits (111), Expect = 5e-06
Identities = 29/99 (29%), Positives = 49/99 (49%), Gaps = 1/99 (1%)
Frame = +2
Query: 388 GITCSEGLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQI-PNLSSLTML 446
GI G +++ + LSG + + + + L ++L N LSG + P L L L
Sbjct: 38 GIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELGHLKSL 217
Query: 447 ETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIP 485
+++ L N SG++P+S + +L + L N L G IP
Sbjct: 218 KSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIP 334
Score = 39.3 bits (90), Expect = 0.001
Identities = 24/65 (36%), Positives = 33/65 (49%), Gaps = 2/65 (3%)
Frame = +2
Query: 426 IWLG--NNSLSGQIPNLSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQ 483
++LG NN G P + +LT L SGEIP+ LG + L +FLQ N L+G
Sbjct: 5 LYLGYYNNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGS 184
Query: 484 IPANL 488
+ L
Sbjct: 185 LTPEL 199
Score = 29.3 bits (64), Expect = 1.5
Identities = 14/37 (37%), Positives = 20/37 (53%)
Frame = +2
Query: 454 NQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANLLK 490
N + G IP +GN++ L L+G+IPA L K
Sbjct: 23 NNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGK 133
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,408,533
Number of Sequences: 28460
Number of extensions: 135962
Number of successful extensions: 905
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 774
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 846
length of query: 496
length of database: 4,897,600
effective HSP length: 94
effective length of query: 402
effective length of database: 2,222,360
effective search space: 893388720
effective search space used: 893388720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC137838.4


