

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137838.2 - phase: 0
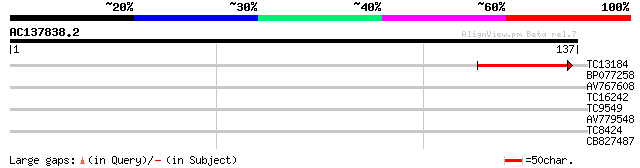
(137 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC13184 43 2e-05
BP077258 28 0.43
AV767608 27 1.6
TC16242 UP|Q8W538 (Q8W538) Ribosomal S15 protein (Fragment), com... 27 1.6
TC9549 weakly similar to UP|Q9S7B6 (Q9S7B6) F1C9.1 protein, part... 26 2.8
AV779548 25 4.7
TC8424 weakly similar to GB|BAC22265.1|24414015|AP003803 arm rep... 25 4.7
CB827487 24 8.1
>TC13184
Length = 538
Score = 43.1 bits (100), Expect = 2e-05
Identities = 18/23 (78%), Positives = 21/23 (91%)
Frame = +3
Query: 114 PLPFEHEVEKGSAFALGLLAVKV 136
PLPFEHEV+KGSAF +GLL +KV
Sbjct: 123 PLPFEHEVQKGSAFGVGLLGIKV 191
>BP077258
Length = 442
Score = 28.5 bits (62), Expect = 0.43
Identities = 20/93 (21%), Positives = 40/93 (42%)
Frame = +3
Query: 14 RKGQKRKLDEELPEDRQISSAPPTADERAALLVEVANQVTVLESTFTWNEADRAAAKRAT 73
+KG K + D +LP+ +++ PP A+ + + L+ T + + T
Sbjct: 99 QKGLKNQ*D*QLPKQPNLNTKPPKANRQTCKNPNEPKPIKTLKELGTNQDLKLLRPQGTT 278
Query: 74 HALADLAKNEEVVNVIVEGGAIPALIKHLQAPP 106
+ ++ + +VEG I A+I H + P
Sbjct: 279 STPQRRLVHIDLGHYLVEGNIICAVIPHATSLP 377
>AV767608
Length = 398
Score = 26.6 bits (57), Expect = 1.6
Identities = 16/37 (43%), Positives = 21/37 (56%)
Frame = +1
Query: 97 ALIKHLQAPPVTDCVQKPLPFEHEVEKGSAFALGLLA 133
ALI+ LQ P+ C + F E+E SA +GLLA
Sbjct: 211 ALIQGLQPIPICFCFLISITFLGELECNSARKVGLLA 321
>TC16242 UP|Q8W538 (Q8W538) Ribosomal S15 protein (Fragment), complete
Length = 581
Score = 26.6 bits (57), Expect = 1.6
Identities = 10/19 (52%), Positives = 13/19 (67%)
Frame = -3
Query: 92 GGAIPALIKHLQAPPVTDC 110
G ++PAL HL PPV+ C
Sbjct: 297 GESLPALASHLVEPPVSPC 241
>TC9549 weakly similar to UP|Q9S7B6 (Q9S7B6) F1C9.1 protein, partial
(83%)
Length = 1560
Score = 25.8 bits (55), Expect = 2.8
Identities = 13/38 (34%), Positives = 21/38 (55%)
Frame = +2
Query: 9 HCLSERKGQKRKLDEELPEDRQISSAPPTADERAALLV 46
HC+ ER G K+ + +++ PT++E AAL V
Sbjct: 41 HCVCER-GDKQGFESRSSHTETMTTVVPTSEEDAALSV 151
>AV779548
Length = 503
Score = 25.0 bits (53), Expect = 4.7
Identities = 11/29 (37%), Positives = 18/29 (61%)
Frame = +2
Query: 88 VIVEGGAIPALIKHLQAPPVTDCVQKPLP 116
V+ EG A+PA++ + +D V+KP P
Sbjct: 404 VLSEGNAVPAIVATTRPEISSDDVRKPNP 490
>TC8424 weakly similar to GB|BAC22265.1|24414015|AP003803 arm repeat
containing protein homolog-like {Oryza sativa (japonica
cultivar-group);} , partial (49%)
Length = 1586
Score = 25.0 bits (53), Expect = 4.7
Identities = 14/43 (32%), Positives = 23/43 (52%)
Frame = +3
Query: 57 STFTWNEADRAAAKRATHALADLAKNEEVVNVIVEGGAIPALI 99
S F+ +D + A AL +L+ N+E +I GA+ +LI
Sbjct: 372 SDFSACSSDPWTQEHAVTALLNLSLNDENKKLITNAGAVKSLI 500
>CB827487
Length = 569
Score = 24.3 bits (51), Expect = 8.1
Identities = 11/24 (45%), Positives = 15/24 (61%)
Frame = +3
Query: 74 HALADLAKNEEVVNVIVEGGAIPA 97
+ LA L E + N+IV+G IPA
Sbjct: 414 YPLAPLIPRETIKNIIVDGYEIPA 485
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.131 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,033,773
Number of Sequences: 28460
Number of extensions: 19522
Number of successful extensions: 115
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 115
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 115
length of query: 137
length of database: 4,897,600
effective HSP length: 81
effective length of query: 56
effective length of database: 2,592,340
effective search space: 145171040
effective search space used: 145171040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 50 (23.9 bits)
Medicago: description of AC137838.2


