

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137831.4 + phase: 0 /partial
(420 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
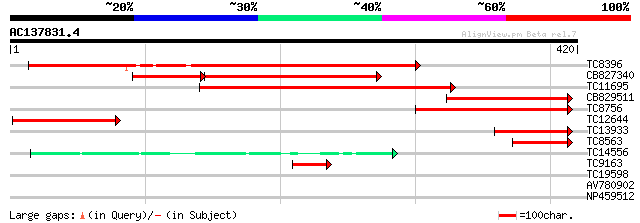
Score E
Sequences producing significant alignments: (bits) Value
TC8396 homologue to UP|Q84UT1 (Q84UT1) ADP-glucose pyrophosphory... 334 1e-92
CB827340 178 7e-65
TC11695 homologue to UP|Q9AT08 (Q9AT08) ADP-glucose pyrophosphor... 241 2e-64
CB829511 141 2e-34
TC8756 homologue to UP|Q9AT06 (Q9AT06) ADP-glucose pyrophosphory... 137 3e-33
TC12644 similar to UP|Q9AT07 (Q9AT07) ADP-glucose pyrophosphoryl... 126 6e-30
TC13933 similar to GB|BAA76362.1|4586350|AB022891 glucose-1-phos... 88 3e-18
TC8563 similar to UP|O22631 (O22631) ADP-glucose pyrophosphoryla... 67 6e-12
TC14556 homologue to UP|Q9ZTW5 (Q9ZTW5) GDP-mannose pyrophosphor... 46 1e-05
TC9163 similar to UP|Q9AT45 (Q9AT45) ADP-glucose pyrophosphoryla... 40 7e-04
TC19598 similar to GB|AAM70562.1|21700877|AY124853 At1g74910/F9E... 30 0.75
AV780902 28 3.7
NP459512 PSI P700 apoprotein A1 [Lotus japonicus] 27 8.2
>TC8396 homologue to UP|Q84UT1 (Q84UT1) ADP-glucose pyrophosphorylase small
subunit PvAGPS1 , partial (67%)
Length = 1223
Score = 334 bits (857), Expect = 1e-92
Identities = 172/294 (58%), Positives = 220/294 (74%), Gaps = 4/294 (1%)
Frame = +1
Query: 15 KTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEINKIYVLTQFNS 74
++V IILGGGAGTRL+PLT+KRAKPAVP G YRL+DIP+SNC+NS ++KIYVLTQFNS
Sbjct: 361 RSVLGIILGGGAGTRLYPLTKKRAKPAVPLGANYRLIDIPVSNCLNSNVSKIYVLTQFNS 540
Query: 75 QSLNRHIARTY--NLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRRFLWLFEDA 132
SLNRH++R Y N+GG N G F+EVLAA Q+ WFQGTADAVR++LWLFE+
Sbjct: 541 ASLNRHLSRAYASNMGGYKNEG--FVEVLAAQQS--PENPNWFQGTADAVRQYLWLFEE- 705
Query: 133 EHRNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDERGR 192
N+ LVL GD LYRMDY + +Q H + ADI+V+ LP+D RA+ FGL+K+DE GR
Sbjct: 706 --HNVLEFLVLAGDHLYRMDYEKFIQAHRETDADITVAALPMDEQRATAFGLMKIDEEGR 879
Query: 193 IHQFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKLLRSCYPNA 252
I +F EKPKG+ L++M VDT++ GL + A++ P+IASMGIYV +V+ LLR +P A
Sbjct: 880 IIEFAEKPKGEQLKAMKVDTTILGLDDERAKEMPFIASMGIYVVSKNVMLDLLREKFPGA 1059
Query: 253 NDFGSEVIPMAAK-DFKVQACLFNGYWEDIGTIKSFFDANLALMDKP-PKFQLY 304
NDFGSEVIP A +VQA L++GYWEDIGTI++F++ANL + KP P F Y
Sbjct: 1060NDFGSEVIPGATSIGMRVQAYLYDGYWEDIGTIEAFYNANLGITKKPVPDFSFY 1221
>CB827340
Length = 555
Score = 178 bits (451), Expect(2) = 7e-65
Identities = 86/131 (65%), Positives = 106/131 (80%)
Frame = +2
Query: 145 GDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDERGRIHQFMEKPKGDL 204
GD LYRMDYM+ VQKHI++ ADI+VSCLP+D SRASDFGL+K+D+ GRI QF EKPKG
Sbjct: 161 GDHLYRMDYMDFVQKHIDTNADITVSCLPMDDSRASDFGLMKIDKTGRIVQFAEKPKGSD 340
Query: 205 LRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKLLRSCYPNANDFGSEVIPMAA 264
L++MHVDT++ GLS +EA+ PYIASMG+YVF+ +VL KLLR + + NDFGSE+IP A
Sbjct: 341 LKAMHVDTTLLGLSPEEAKNNPYIASMGVYVFRTEVLLKLLRWSHSSCNDFGSEIIPSAV 520
Query: 265 KDFKVQACLFN 275
D VQA LFN
Sbjct: 521 SDHNVQAYLFN 553
Score = 86.3 bits (212), Expect(2) = 7e-65
Identities = 37/54 (68%), Positives = 48/54 (88%)
Frame = +3
Query: 92 NCGGSFIEVLAATQTLGESGNKWFQGTADAVRRFLWLFEDAEHRNIENILVLCG 145
N G F+EVLAATQT GE+G KWFQGTADAVR+F+W+FEDA+++N+E+IL+L G
Sbjct: 3 NFGNGFVEVLAATQTPGEAGKKWFQGTADAVRQFIWVFEDAKNKNVEHILILLG 164
>TC11695 homologue to UP|Q9AT08 (Q9AT08) ADP-glucose pyrophosphorylase
(Glucose-1-phosphate adenylyltransferase) (ADP-glucose
synthase) , partial (36%)
Length = 575
Score = 241 bits (615), Expect = 2e-64
Identities = 116/190 (61%), Positives = 148/190 (77%)
Frame = +1
Query: 141 LVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDERGRIHQFMEKP 200
L+L GD LYRMDYM+ VQ H S ADI++SCLP+D SRASDFGL+K+D +GR+ F EKP
Sbjct: 4 LILSGDHLYRMDYMDFVQNHRESGADITLSCLPMDDSRASDFGLMKIDNKGRVLSFSEKP 183
Query: 201 KGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKLLRSCYPNANDFGSEVI 260
KG L++M VDT+V GLS EA K PYIASMG+YVFK ++L LLR +P ANDFGSEVI
Sbjct: 184 KGADLKAMQVDTTVLGLSKDEAIKKPYIASMGVYVFKKEILLNLLRWRFPTANDFGSEVI 363
Query: 261 PMAAKDFKVQACLFNGYWEDIGTIKSFFDANLALMDKPPKFQLYDQSKPIFTCPRFLPPT 320
P +A +F ++A LFN YWEDIGTI+SFF+ANLAL + P KF YD +KP++T R LPP+
Sbjct: 364 PASAAEFYMKAYLFNDYWEDIGTIRSFFEANLALTEHPSKFSFYDAAKPMYTSRRNLPPS 543
Query: 321 KLEKCQVVNS 330
K++ ++V+S
Sbjct: 544 KIDNSKIVDS 573
>CB829511
Length = 572
Score = 141 bits (355), Expect = 2e-34
Identities = 66/94 (70%), Positives = 82/94 (87%)
Frame = +2
Query: 324 KCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQLKDTMMMGADYYETEAEIASLLSA 383
KC+VV++++S GCFLREC V+HSIVG RSRL+ GV L+DT+MMGADYY+TE+EIASLL+
Sbjct: 2 KCRVVDAIISHGCFLRECTVQHSIVGERSRLDYGVDLQDTIMMGADYYQTESEIASLLAE 181
Query: 384 GDVPIGIGKNTKIMKCIIDKNARIGNNVTIANKE 417
G VPIGIG+NTKI CIIDKNA+IG +V I NK+
Sbjct: 182 GKVPIGIGRNTKIRNCIIDKNAKIGKDVIITNKD 283
>TC8756 homologue to UP|Q9AT06 (Q9AT06) ADP-glucose pyrophosphorylase small
subunit CagpS1 (Glucose-1-phosphate
adenylyltransferase) (ADP-glucose synthase) , partial
(29%)
Length = 771
Score = 137 bits (345), Expect = 3e-33
Identities = 59/117 (50%), Positives = 87/117 (73%)
Frame = +3
Query: 301 FQLYDQSKPIFTCPRFLPPTKLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQL 360
F YD+S PI+T PR+LPP+K+ + +S++ +GC ++ CK+ HS+VG+RS ++ G +
Sbjct: 6 FSFYDRSSPIYTQPRYLPPSKMLDADITDSVIGEGCVIKNCKIHHSVVGLRSCVSEGAII 185
Query: 361 KDTMMMGADYYETEAEIASLLSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTIANKE 417
+DT++MGADYYET+A+ L + G VPIGIG+N+ I + IIDKNARIG NV I N +
Sbjct: 186 EDTLLMGADYYETDADKRFLAAKGSVPIGIGRNSHIKRAIIDKNARIGENVKIINTD 356
>TC12644 similar to UP|Q9AT07 (Q9AT07) ADP-glucose pyrophosphorylase large
subunit CagpL2 (Glucose-1-phosphate
adenylyltransferase) (ADP-glucose synthase) , partial
(17%)
Length = 833
Score = 126 bits (317), Expect = 6e-30
Identities = 60/80 (75%), Positives = 69/80 (86%)
Frame = +2
Query: 3 QSPIFAGQEANPKTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSE 62
+S F +A+PK+VASII GGGAGTRLFPLT +RAKPAVP GGCYRL+DIPMSNCINS
Sbjct: 593 ESHTFETSKADPKSVASIIXGGGAGTRLFPLTSRRAKPAVPXGGCYRLIDIPMSNCINSG 772
Query: 63 INKIYVLTQFNSQSLNRHIA 82
I KI++LTQFNS SLNRH+A
Sbjct: 773 IRKIFILTQFNSFSLNRHLA 832
>TC13933 similar to GB|BAA76362.1|4586350|AB022891 glucose-1-phosphate
adenylyltransferase {Arabidopsis thaliana;} , partial
(17%)
Length = 560
Score = 87.8 bits (216), Expect = 3e-18
Identities = 40/58 (68%), Positives = 50/58 (85%)
Frame = +2
Query: 360 LKDTMMMGADYYETEAEIASLLSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTIANKE 417
++DT+M+GAD+YETE E A+LL+ G VP+GIG+NTKI +CIIDKNARIG NV IAN E
Sbjct: 53 VQDTVMLGADFYETETEAAALLAEGGVPVGIGENTKIRECIIDKNARIGKNVIIANSE 226
>TC8563 similar to UP|O22631 (O22631) ADP-glucose pyrophosphorylase large
subunit , partial (15%)
Length = 917
Score = 67.0 bits (162), Expect = 6e-12
Identities = 31/45 (68%), Positives = 37/45 (81%)
Frame = -1
Query: 373 TEAEIASLLSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTIANKE 417
TE EIASL++ G VPIG+G+NTKI CIIDKNA+IG NV IAN +
Sbjct: 917 TETEIASLVAEGKVPIGVGENTKIRNCIIDKNAKIGRNVVIANTD 783
>TC14556 homologue to UP|Q9ZTW5 (Q9ZTW5) GDP-mannose pyrophosphorylase,
complete
Length = 1521
Score = 46.2 bits (108), Expect = 1e-05
Identities = 60/274 (21%), Positives = 109/274 (38%), Gaps = 2/274 (0%)
Frame = +3
Query: 16 TVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEINKIYVLTQFNSQ 75
T+ ++IL GG GTRL PLT KP V F ++ + + + ++ + + +
Sbjct: 156 TMKALILVGGFGTRLRPLTLSFPKPLVDFANKPMILH-QIEALKAAGVTEVVLAINYQPE 332
Query: 76 SLNRHIAR-TYNLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRRFLWLFEDAEH 134
+ + LG ++C E L L + +K G+
Sbjct: 333 VMLNFLKDFEAKLGIKISCSQE-TEPLGTAGPLALARDKLIDGSG--------------- 464
Query: 135 RNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDER-GRI 193
E VL D + E+++ H + + + VD S +G+V ++E G++
Sbjct: 465 ---EPFFVLNSDVISEYPLEEMIEFHKSHGGEAFIMVTKVD--EPSKYGVVVMEESTGQV 629
Query: 194 HQFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKLLRSCYPNAN 253
+F+EKPK + V + + GIY+ VL ++ P +
Sbjct: 630 EKFVEKPK------LFVGNKI---------------NAGIYLLNPSVLDRI--ELRPTSI 740
Query: 254 DFGSEVIPMAAKDFKVQACLFNGYWEDIGTIKSF 287
+ EV P A + K+ A + G+W DIG + +
Sbjct: 741 E--KEVFPKIAAEQKLYAMVLPGFWMDIGQPRDY 836
>TC9163 similar to UP|Q9AT45 (Q9AT45) ADP-glucose pyrophosphorylase large
subunit (Glucose-1-phosphate adenylyltransferase)
(ADP-glucose synthase) , partial (5%)
Length = 663
Score = 40.0 bits (92), Expect = 7e-04
Identities = 19/30 (63%), Positives = 24/30 (79%), Gaps = 1/30 (3%)
Frame = +2
Query: 210 VDTSVFGLSAQEARKFP-YIASMGIYVFKL 238
VDT+V GLS +EA P YIASMG+Y+FK+
Sbjct: 107 VDTTVLGLSQEEAENKPSYIASMGVYIFKM 196
>TC19598 similar to GB|AAM70562.1|21700877|AY124853 At1g74910/F9E10_24
{Arabidopsis thaliana;}, partial (27%)
Length = 495
Score = 30.0 bits (66), Expect = 0.75
Identities = 20/51 (39%), Positives = 27/51 (52%), Gaps = 1/51 (1%)
Frame = +2
Query: 9 GQEANPKTVASIILGGGA-GTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNC 58
G + + VA I++GG GTR PL+ AKP P G +V P+S C
Sbjct: 146 GIAEDERVVAVIMVGGPTKGTRFRPLSLNIAKPLFPLAG-QPMVHHPISAC 295
>AV780902
Length = 506
Score = 27.7 bits (60), Expect = 3.7
Identities = 13/32 (40%), Positives = 18/32 (55%)
Frame = -2
Query: 247 SCYPNANDFGSEVIPMAAKDFKVQACLFNGYW 278
SC NAND ++ ++FK+ CLFN W
Sbjct: 283 SCLVNAND----LVH*VDRNFKITGCLFNMEW 200
>NP459512 PSI P700 apoprotein A1 [Lotus japonicus]
Length = 2253
Score = 26.6 bits (57), Expect = 8.2
Identities = 14/39 (35%), Positives = 19/39 (47%)
Frame = +1
Query: 115 FQGTADAVRRFLWLFEDAEHRNIENILVLCGDQLYRMDY 153
F+G D V LWL + A H IL L +YR ++
Sbjct: 823 FRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNW 939
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.139 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,120,924
Number of Sequences: 28460
Number of extensions: 98860
Number of successful extensions: 431
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 423
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 427
length of query: 420
length of database: 4,897,600
effective HSP length: 93
effective length of query: 327
effective length of database: 2,250,820
effective search space: 736018140
effective search space used: 736018140
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC137831.4


