

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137825.10 - phase: 0 /pseudo
(527 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
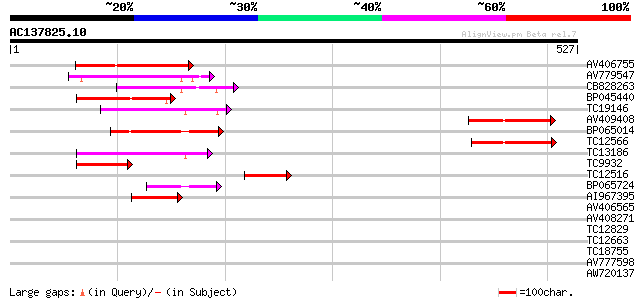
Score E
Sequences producing significant alignments: (bits) Value
AV406755 115 1e-26
AV779547 99 1e-21
CB828263 89 2e-18
BP045440 87 5e-18
TC19146 similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-contain... 84 6e-17
AV409408 84 7e-17
BP065014 81 5e-16
TC12566 similar to UP|Q84XI1 (Q84XI1) RCa1 (Fragment), partial (... 77 5e-15
TC13186 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candidat... 68 3e-12
TC9932 weakly similar to UP|Q93YA7 (Q93YA7) Resistance gene-like... 58 4e-09
TC12516 similar to UP|Q9M5V7 (Q9M5V7) Disease resistance-like pr... 55 2e-08
BP065724 50 7e-07
AI967395 48 3e-06
AV406565 39 0.002
AV408271 39 0.003
TC12829 37 0.006
TC12663 weakly similar to UP|Q84ZV8 (Q84ZV8) R 3 protein, partia... 35 0.008
TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fr... 34 0.052
AV777598 30 0.46
AW720137 29 2.2
>AV406755
Length = 425
Score = 115 bits (289), Expect = 1e-26
Identities = 57/110 (51%), Positives = 80/110 (71%)
Frame = +3
Query: 62 SYEVFLSYRGEDTRASFTAHLNASLLNAGINVFKDDDSIYKGARISKSLPEAIEQSRIAV 121
SY+VFLS+RGE++R SFT+HL +L NAGI VF D++ + +G IS SL +AIE SRIA+
Sbjct: 81 SYDVFLSFRGEESRRSFTSHLYTALKNAGIKVFMDNE-LQRGEDISSSLLKAIEDSRIAI 257
Query: 122 VVFSKHYADSKWCLNELVKIMKCHRAIRQIVLPVFYDTQGEGHYKEESSL 171
++FS +Y SKWCL+EL KI++C R I Q V+PVFY+ K+ ++
Sbjct: 258 IIFSTNYTGSKWCLDELEKIIECQRTIGQEVMPVFYNVDPSDIRKQRGTV 407
>AV779547
Length = 556
Score = 99.4 bits (246), Expect = 1e-21
Identities = 66/155 (42%), Positives = 83/155 (52%), Gaps = 19/155 (12%)
Frame = +1
Query: 55 SPLKIFRSYEV----FLSYRGEDTRASFTAHLNASLLNAGINVFKDDDSIYKGARISKSL 110
+P ++F S ++ +S+RGEDTR +FT HL +L G FKDD + KG IS L
Sbjct: 61 NPKQLFNSDDMEI*CVVSFRGEDTRNNFTDHLLGALYGKGFVTFKDDTMLRKGKNISTEL 240
Query: 111 PEAIEQSRIAVVVFSKHYADSKWCLNELVKIMKCHRAIRQIVLPVFYD-----------T 159
+AIE S+I +VVFSK+YA S WCL EL KI RQ VLPV D
Sbjct: 241 IQAIEGSQILIVVFSKYYASSTWCLQELAKIADGIVGKRQTVLPVVCDVTPSEVRKQSGN 420
Query: 160 QGEGHYKEE----SSLRMLRRNWTTALHEAAGLAG 190
GE K E L M++R W AL E A L+G
Sbjct: 421 YGEAFLKHEERFKEDLEMVQR-WRKALAEVADLSG 522
>CB828263
Length = 570
Score = 88.6 bits (218), Expect = 2e-18
Identities = 52/131 (39%), Positives = 77/131 (58%), Gaps = 18/131 (13%)
Frame = +2
Query: 100 IYKGARISKSLPEAIEQSRIAVVVFSKHYADSKWCLNELVKIMKCHRAIRQIVLPVFYD- 158
+ +G IS SL AIE+++++V+VFSK+Y +SKWCL+ELVKI++C R QIVLPVFYD
Sbjct: 8 LQRGDEISSSLLRAIEEAKLSVIVFSKNYGNSKWCLDELVKILECKRTRGQIVLPVFYDI 187
Query: 159 ----------TQGEGHYKEESSLRMLRRNWTTALHEAAGLAG-------NESEAIKDIVE 201
T E K ++ + W AL EAA L+G ESE I+ I +
Sbjct: 188 DPSHVRNQTGTYAEAFVKHGQVDKV--QKWREALREAANLSGWDCSVNRMESEVIEKIAK 361
Query: 202 KIVRLLDKTHL 212
++ L++ ++
Sbjct: 362 DVLEKLNRVYV 394
>BP045440
Length = 535
Score = 87.4 bits (215), Expect = 5e-18
Identities = 44/94 (46%), Positives = 62/94 (65%), Gaps = 2/94 (2%)
Frame = -3
Query: 63 YEVFLSYRGEDTRASFTAHLNASLLNAGINVFKDDDSIYKGARISKSLPEAIEQSRIAVV 122
Y++F+S+RG DTR FT L L G FKDD+S+ G+ I K L + IE+SR A++
Sbjct: 527 YQIFISFRGMDTRDDFTKPLYQGLCREGFVTFKDDESLEGGSPIEKLLDD-IEESRFAII 351
Query: 123 VFSKHYADSKWCLNELVKIMKC--HRAIRQIVLP 154
+ SK+YA+S+WCL EL KI++C Q+VLP
Sbjct: 350 ILSKNYAESEWCLKELSKILECKDQEGKNQLVLP 249
>TC19146 similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-containing protein
TSDC, partial (12%)
Length = 701
Score = 84.0 bits (206), Expect = 6e-17
Identities = 50/143 (34%), Positives = 74/143 (50%), Gaps = 21/143 (14%)
Frame = +1
Query: 85 SLLNAGINVFKDDDSIYKGARISKSLPEAIEQSRIAVVVFSKHYADSKWCLNELVKIMKC 144
+L G F DD+ + G I+KSL +A E+SR++++VFS++Y S WCL+ELVKI++C
Sbjct: 10 ALRREGFKTFMDDEGLVGGNEIAKSLLDASEKSRLSIIVFSENYGYSSWCLDELVKIVEC 189
Query: 145 HRAIRQIVLPVFYDTQGE--------------GHYKEESSLRMLRRNWTTALHEAAGLAG 190
+Q+V P+FY + GH K + W +AL A L G
Sbjct: 190 METKKQLVWPIFYKVEKSDVTSQENSYEHAMIGHEKNYGKDSDEVKKWRSALSAMAKLEG 369
Query: 191 N-------ESEAIKDIVEKIVRL 206
+ E E IK IVEK + +
Sbjct: 370 HNVKENKYEYEFIKKIVEKAIAI 438
>AV409408
Length = 430
Score = 83.6 bits (205), Expect = 7e-17
Identities = 37/81 (45%), Positives = 59/81 (72%)
Frame = +2
Query: 427 VTEWENVLEKLKRIPNGQVQKKLKISYDALNDDTQEDIFLNIACFFIGLVRNNVIHILNG 486
+++WE+ L K+K++P+ + KL+ISYD L D+ + +FL+IACFF G ++ VI +L
Sbjct: 56 LSDWEDALIKIKKVPHDDILNKLRISYDMLEDE-HKTLFLDIACFFKGWYKHKVIQLLGS 232
Query: 487 CELYAEIGINVLVERSLVTID 507
C L+ +GINVL+E+SL+T D
Sbjct: 233 CGLHPTVGINVLIEKSLLTFD 295
>BP065014
Length = 498
Score = 80.9 bits (198), Expect = 5e-16
Identities = 43/105 (40%), Positives = 67/105 (62%)
Frame = -2
Query: 94 FKDDDSIYKGARISKSLPEAIEQSRIAVVVFSKHYADSKWCLNELVKIMKCHRAIRQIVL 153
FKDD S+ GA I + L AIE+SR+A+VV S+++ADSKWCL ELVKI+ C + + +VL
Sbjct: 497 FKDDLSLEDGAPIEELLG-AIEESRVAIVVLSENFADSKWCLKELVKILDCRKKKKPLVL 321
Query: 154 PVFYDTQGEGHYKEESSLRMLRRNWTTALHEAAGLAGNESEAIKD 198
P+FY E +++R L+ + A+ E G +S+ +++
Sbjct: 320 PIFYKV-------EPTNIRHLKGRYGEAMAEHEKKLGKDSQTVRE 207
>TC12566 similar to UP|Q84XI1 (Q84XI1) RCa1 (Fragment), partial (28%)
Length = 918
Score = 77.4 bits (189), Expect = 5e-15
Identities = 36/79 (45%), Positives = 58/79 (72%)
Frame = +1
Query: 430 WENVLEKLKRIPNGQVQKKLKISYDALNDDTQEDIFLNIACFFIGLVRNNVIHILNGCEL 489
WE+ L KL+++P+ ++ LK+SYD L+DD Q+DIFL+IACF++G + +V L+ C
Sbjct: 196 WESQLVKLEKLPDPEIINVLKLSYDGLDDD-QKDIFLDIACFYLGHLEESVAQTLDSCGF 372
Query: 490 YAEIGINVLVERSLVTIDE 508
+A+IG+ VL +R L++I E
Sbjct: 373 FADIGMEVLKDRCLISILE 429
>TC13186 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candidate
resistance protein KR1, partial (6%)
Length = 586
Score = 68.2 bits (165), Expect = 3e-12
Identities = 47/140 (33%), Positives = 66/140 (46%), Gaps = 14/140 (10%)
Frame = +1
Query: 63 YEVFLSYRGEDTRASFTAHLNASLLNAGINVFKDDDSIYKGARISKSLPEAIEQSRIAVV 122
Y+VF+S DTR F+ L +L + GI F DD +S +AIE SRIA+V
Sbjct: 103 YDVFISLITSDTRFGFSGDLYTALSDKGILTFIDDGLPRPDDIMSTVNSKAIESSRIAIV 282
Query: 123 VFSKHYADSKWCLNELVKIMKCHRAIRQIVLPVFYDTQGE--------------GHYKEE 168
V S++YA S +CL+EL+ I+K ++ PVFY + H +
Sbjct: 283 VISENYASSSFCLDELMYILKRREEKGLLIWPVFYQVEPSDVRYQKGGFGKAFASHEERF 462
Query: 169 SSLRMLRRNWTTALHEAAGL 188
S R W AL + A L
Sbjct: 463 GSDSAKVRKWRNALKQVADL 522
>TC9932 weakly similar to UP|Q93YA7 (Q93YA7) Resistance gene-like, partial
(5%)
Length = 689
Score = 57.8 bits (138), Expect = 4e-09
Identities = 24/52 (46%), Positives = 38/52 (72%)
Frame = -2
Query: 63 YEVFLSYRGEDTRASFTAHLNASLLNAGINVFKDDDSIYKGARISKSLPEAI 114
Y+VF+++RG+DTR SF +HL +L NAG+NVF DD+++ +G + L + I
Sbjct: 202 YDVFINFRGKDTRHSFVSHLYTALSNAGVNVFLDDENLERGLELEPELQKLI 47
>TC12516 similar to UP|Q9M5V7 (Q9M5V7) Disease resistance-like protein
(Fragment), partial (36%)
Length = 859
Score = 55.5 bits (132), Expect = 2e-08
Identities = 23/44 (52%), Positives = 36/44 (81%)
Frame = +3
Query: 219 VGVEPRVENMIQLLDEKSKDVLLLGMWGMGGIGKTTIAKAIYNK 262
+GV+ + ++ LL ++SKDV ++G+WGMGGIGKTTIA+ I+N+
Sbjct: 549 IGVDKSIAHLESLLCQESKDVRVIGIWGMGGIGKTTIAEEIFNR 680
>BP065724
Length = 495
Score = 50.4 bits (119), Expect = 7e-07
Identities = 26/70 (37%), Positives = 39/70 (55%)
Frame = -1
Query: 128 YADSKWCLNELVKIMKCHRAIRQIVLPVFYDTQGEGHYKEESSLRMLRRNWTTALHEAAG 187
YADS WCL E+VKI++C + Q+V P+FY E S +R R ++ A+ +
Sbjct: 495 YADSSWCLEEVVKILECKQKNDQLVWPIFYKV-------EPSDVRHQRNSYEKAMIKQET 337
Query: 188 LAGNESEAIK 197
G +SE +K
Sbjct: 336 RYGKDSEKVK 307
>AI967395
Length = 205
Score = 48.1 bits (113), Expect = 3e-06
Identities = 23/47 (48%), Positives = 32/47 (67%)
Frame = +3
Query: 114 IEQSRIAVVVFSKHYADSKWCLNELVKIMKCHRAIRQIVLPVFYDTQ 160
I++SR+ +VVFS+HYA S CLNELV I+ Q+V PV+Y +
Sbjct: 3 IQESRLIIVVFSEHYAVSASCLNELVNIVDVMIMNNQLVWPVYYGVE 143
>AV406565
Length = 207
Score = 38.9 bits (89), Expect = 0.002
Identities = 17/38 (44%), Positives = 25/38 (65%)
Frame = +2
Query: 61 RSYEVFLSYRGEDTRASFTAHLNASLLNAGINVFKDDD 98
+ ++VFLS+RGEDTR + T HL A+L + + D D
Sbjct: 71 QKHDVFLSFRGEDTRYTCTGHLQATLTRLQVKTYIDYD 184
>AV408271
Length = 409
Score = 38.5 bits (88), Expect = 0.003
Identities = 17/35 (48%), Positives = 22/35 (62%)
Frame = +1
Query: 63 YEVFLSYRGEDTRASFTAHLNASLLNAGINVFKDD 97
Y+VF+S+RGED R F +HL + IN F DD
Sbjct: 280 YDVFVSFRGEDIRDGFLSHLADAFRQKKINAFMDD 384
>TC12829
Length = 448
Score = 37.4 bits (85), Expect = 0.006
Identities = 20/57 (35%), Positives = 32/57 (56%), Gaps = 2/57 (3%)
Frame = +3
Query: 200 VEKIVRLLDKTHLLVANNPVGVEPRVENMIQLLDE--KSKDVLLLGMWGMGGIGKTT 254
V+ ++ L +N VG++P +E + LL + +D +LG+W GGIGKTT
Sbjct: 54 VQARIKTLGHKFSRFSNGLVGIQPHIEEI*NLLKLS*EDEDFRVLGIWRTGGIGKTT 224
>TC12663 weakly similar to UP|Q84ZV8 (Q84ZV8) R 3 protein, partial (4%)
Length = 494
Score = 35.0 bits (79), Expect(2) = 0.008
Identities = 16/41 (39%), Positives = 26/41 (63%)
Frame = +3
Query: 62 SYEVFLSYRGEDTRASFTAHLNASLLNAGINVFKDDDSIYK 102
++ VFL++RG DT F + L SLL+ GI F +D+ + +
Sbjct: 24 TFNVFLNFRGSDTCHGFISFLCKSLLDKGIYTFINDEELQR 146
Score = 20.8 bits (42), Expect(2) = 0.008
Identities = 9/16 (56%), Positives = 14/16 (87%)
Frame = +2
Query: 106 ISKSLPEAIEQSRIAV 121
I+ SL +AIE++RIA+
Sbjct: 155 ITPSLVKAIERTRIAI 202
>TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fragment),
complete
Length = 880
Score = 34.3 bits (77), Expect = 0.052
Identities = 19/56 (33%), Positives = 32/56 (56%)
Frame = +3
Query: 206 LLDKTHLLVANNPVGVEPRVENMIQLLDEKSKDVLLLGMWGMGGIGKTTIAKAIYN 261
LLD T L VG++ R + ++ L + ++ + GMGG+GKTT+ K +Y+
Sbjct: 177 LLDNTDL------VGIDRRKKKLMGCLIKPCPVRKVISVTGMGGMGKTTLVKQVYD 326
>AV777598
Length = 371
Score = 30.0 bits (66), Expect(2) = 0.46
Identities = 13/21 (61%), Positives = 17/21 (80%)
Frame = +3
Query: 62 SYEVFLSYRGEDTRASFTAHL 82
+Y+VFLS+ G DTR SFT +L
Sbjct: 306 TYDVFLSFCGVDTRFSFTGNL 368
Score = 19.6 bits (39), Expect(2) = 0.46
Identities = 6/7 (85%), Positives = 6/7 (85%)
Frame = +2
Query: 13 IACHLFC 19
I CHLFC
Sbjct: 263 IECHLFC 283
>AW720137
Length = 563
Score = 28.9 bits (63), Expect = 2.2
Identities = 21/62 (33%), Positives = 31/62 (49%), Gaps = 6/62 (9%)
Frame = +3
Query: 7 RHHIHNIACHL------FCLVCRSSTISSSIPSHPSMSSFYNEDSFSTDCRSRKSPLKIF 60
+HH HN + ++ SS+ SSS+PSHP SF N ++S R + S + F
Sbjct: 375 QHHHHNTTTTIAESDEKHHVLDLSSSSSSSLPSHPFYLSFTNL-TYSVKLRRKMSLSRCF 551
Query: 61 RS 62
S
Sbjct: 552 TS 557
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.349 0.153 0.522
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,091,808
Number of Sequences: 28460
Number of extensions: 121467
Number of successful extensions: 1327
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 1304
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1322
length of query: 527
length of database: 4,897,600
effective HSP length: 94
effective length of query: 433
effective length of database: 2,222,360
effective search space: 962281880
effective search space used: 962281880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC137825.10


