

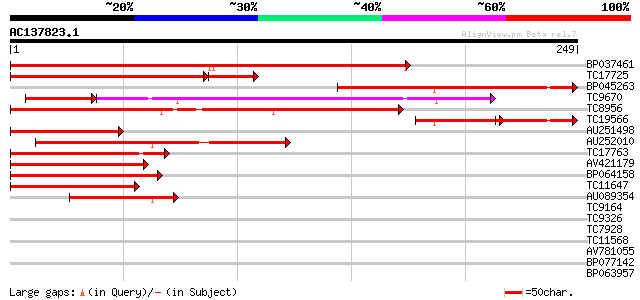
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137823.1 + phase: 0
(249 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP037461 272 5e-74
TC17725 homologue to UP|Q8LLR2 (Q8LLR2) MADS-box protein 2, part... 166 2e-47
BP045263 166 3e-42
TC9670 homologue to UP|O64958 (O64958) CUM1, partial (65%) 104 6e-34
TC8956 similar to UP|Q9ZS28 (Q9ZS28) MADs-box protein, GDEF1, pa... 115 5e-27
TC19566 similar to UP|O82084 (O82084) MADS-box protein 1, partia... 65 4e-21
AU251498 89 7e-19
AU252010 84 3e-17
TC17763 similar to UP|Q7Y1U9 (Q7Y1U9) SVP-like floral repressor,... 81 2e-16
AV421179 78 1e-15
BP064158 74 2e-14
TC11647 similar to UP|Q7X9H7 (Q7X9H7) MADS-box protein AGL62, pa... 70 3e-13
AU089354 46 7e-06
TC9164 weakly similar to UP|Q84MJ0 (Q84MJ0) MADS4 (Fragment), pa... 33 0.046
TC9326 homologue to UP|Q93WX4 (Q93WX4) Suppressor of K+ transpor... 32 0.13
TC7928 similar to UP|Q93YR3 (Q93YR3) HSP associated protein like... 27 4.3
TC11568 similar to UP|Q9AYT8 (Q9AYT8) Mangrin, partial (68%) 26 5.6
AV781055 26 7.4
BP077142 25 9.6
BP063957 25 9.6
>BP037461
Length = 551
Score = 272 bits (695), Expect = 5e-74
Identities = 144/180 (80%), Positives = 162/180 (90%), Gaps = 4/180 (2%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFS RGKLYEFCS+
Sbjct: 8 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 187
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKE-LE-SSYREYLKLKQRFENLQRAQRNLLGEDLGP 118
S+MLKTL+RYQKC+YGA E + +E LE SS +EYLKLK R+E LQR+QRNL+GEDLGP
Sbjct: 188 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 367
Query: 119 LSSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLE--EINS 176
L+SK+LE LERQLDSSLKQ+RST+TQFMLDQL+DLQ KEHML EANRSL +LE ++NS
Sbjct: 368 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLEGYQLNS 547
>TC17725 homologue to UP|Q8LLR2 (Q8LLR2) MADS-box protein 2, partial (45%)
Length = 970
Score = 166 bits (419), Expect(2) = 2e-47
Identities = 81/87 (93%), Positives = 87/87 (99%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALI+FSTRGKLYEFCS+
Sbjct: 276 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIVFSTRGKLYEFCSS 455
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKEL 87
S+M+KTL+RYQKCSYGAVEV+KPAKEL
Sbjct: 456 SSMVKTLERYQKCSYGAVEVNKPAKEL 536
Score = 39.7 bits (91), Expect(2) = 2e-47
Identities = 18/22 (81%), Positives = 20/22 (90%)
Frame = +1
Query: 88 ESSYREYLKLKQRFENLQRAQR 109
+SSYREYLKLK RFE+LQR QR
Sbjct: 655 QSSYREYLKLKGRFESLQRTQR 720
>BP045263
Length = 535
Score = 166 bits (420), Expect = 3e-42
Identities = 82/106 (77%), Positives = 92/106 (86%), Gaps = 1/106 (0%)
Frame = -1
Query: 145 FMLDQLADLQNKEHMLVEANRSLSIKLEEINSRNHYRQSWE-ASDQSMQYEAQQNAHSQS 203
FMLDQL+DLQNKE MLVEANRSLS+KL++INSRNHYR SW+ A D SMQY NAHSQ
Sbjct: 535 FMLDQLSDLQNKEQMLVEANRSLSVKLDDINSRNHYRLSWDQAGDPSMQYGGSPNAHSQG 356
Query: 204 FFQQLECNPTLQIGSDYRYNNVASDQIASTSQAQQQVNGFVPGWML 249
FFQ LECNPTLQIGSDYRY+ VASDQ+A+T+QA Q VNGF+PGWML
Sbjct: 355 FFQPLECNPTLQIGSDYRYHGVASDQMAATTQA-QPVNGFIPGWML 221
>TC9670 homologue to UP|O64958 (O64958) CUM1, partial (65%)
Length = 1060
Score = 104 bits (259), Expect(2) = 6e-34
Identities = 66/183 (36%), Positives = 103/183 (56%), Gaps = 8/183 (4%)
Frame = +3
Query: 39 CDAEVALIIFSTRGKLYEFCSTSNMLKTLDRYQK-CSYGAVEVSKPAKELESSYREYLKL 97
CDAEVALI+FS+RG+LYE+ + S + T+DRY+K CS + S + +E KL
Sbjct: 93 CDAEVALIVFSSRGRLYEYANNS-VKATIDRYKKACSDSSGAGSASEANAQFYQQEADKL 269
Query: 98 KQRFENLQRAQRNLLGEDLGPLSSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKE 157
+ + NLQ R ++GE LG +++K+L+ LE +L+ + ++RS K + + ++ +Q +E
Sbjct: 270 RVQISNLQNNNRQMMGESLGSMNAKELKNLETKLEKGISRIRSKKNELLFAEIEYMQKRE 449
Query: 158 HMLVEANRSLSIKLEEINSRNHYRQSWEA-------SDQSMQYEAQQNAHSQSFFQQLEC 210
L N+ L K+ E + RNH S A S QS Q + QQ S+ +FQ
Sbjct: 450 IDLHNNNQLLRAKIAE-SERNHPNLSILAGSTSNYESMQSQQQQQQQQFDSRGYFQVTGL 626
Query: 211 NPT 213
PT
Sbjct: 627 QPT 635
Score = 55.8 bits (133), Expect(2) = 6e-34
Identities = 27/31 (87%), Positives = 28/31 (90%)
Frame = +1
Query: 8 LKRIENKINRQVTFAKRRNGLLKKAYELSVL 38
+KRIEN NRQVTF KRRNGLLKKAYELSVL
Sbjct: 1 IKRIENTTNRQVTFCKRRNGLLKKAYELSVL 93
>TC8956 similar to UP|Q9ZS28 (Q9ZS28) MADs-box protein, GDEF1, partial
(58%)
Length = 1100
Score = 115 bits (289), Expect = 5e-27
Identities = 70/176 (39%), Positives = 108/176 (60%), Gaps = 3/176 (1%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRG++E+K IEN NRQVT++KRRNG+ KKA+ELSVLCDA+V+LI+FS K++E+ S
Sbjct: 137 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYISP 316
Query: 61 SNMLK-TLDRYQKCSYGAVEVSKPAKELESSYREYLKLKQRFENLQRAQRNLLGE--DLG 117
K +D+YQK + G +++ + E KLK+ L+R R+ LGE D+
Sbjct: 317 GLTTKRIIDQYQK-TLGDIDLWR--SHYEKMLENLKKLKEINNKLRRQIRHRLGEGLDMD 487
Query: 118 PLSSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEE 173
LS + L +LE + SS+ ++R K + + + K L + N +L ++L+E
Sbjct: 488 DLSFQQLRKLEEDMVSSIGKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLELKE 655
>TC19566 similar to UP|O82084 (O82084) MADS-box protein 1, partial (17%)
Length = 531
Score = 65.1 bits (157), Expect(2) = 4e-21
Identities = 30/40 (75%), Positives = 32/40 (80%), Gaps = 1/40 (2%)
Frame = +2
Query: 179 HYRQSWE-ASDQSMQYEAQQNAHSQSFFQQLECNPTLQIG 217
HYRQSW+ A D+SMQY NAHSQ FFQ LECNPTLQIG
Sbjct: 2 HYRQSWDQAGDRSMQYGGSPNAHSQGFFQPLECNPTLQIG 121
Score = 52.0 bits (123), Expect(2) = 4e-21
Identities = 24/36 (66%), Positives = 30/36 (82%)
Frame = +1
Query: 214 LQIGSDYRYNNVASDQIASTSQAQQQVNGFVPGWML 249
L+ SDYRY+ VASDQ+A+T+ A Q VNGF+PGWML
Sbjct: 217 LKFSSDYRYHGVASDQMAATTPA-QPVNGFIPGWML 321
>AU251498
Length = 342
Score = 89.0 bits (219), Expect = 7e-19
Identities = 42/50 (84%), Positives = 47/50 (94%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFST 50
MGRG++E+KRIEN NRQVTF KRRNGLLKKAYELSVLCDAEVALI+FS+
Sbjct: 45 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSS 194
Score = 30.4 bits (67), Expect = 0.30
Identities = 19/74 (25%), Positives = 37/74 (49%), Gaps = 1/74 (1%)
Frame = +1
Query: 2 GRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCSTS 61
G GR+ + + ++ + A+ G +K ++ + RG+LYE+ S +
Sbjct: 49 GGGRLRSRGLRTQLIAR*HSAREETGF*RKLMSYQCCVMLKLPSLSSPARGRLYEY-SNN 225
Query: 62 NMLKTLDRYQK-CS 74
N+ T++RY+K CS
Sbjct: 226 NIRSTIERYKKACS 267
>AU252010
Length = 328
Score = 83.6 bits (205), Expect = 3e-17
Identities = 47/113 (41%), Positives = 75/113 (65%), Gaps = 1/113 (0%)
Frame = +1
Query: 12 ENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCSTS-NMLKTLDRY 70
EN NRQVT++KR+NG+LKKA E++VLCDA+V+LIIF+ GK++++ S S ++ L+RY
Sbjct: 1 ENSSNRQVTYSKRKNGILKKAKEITVLCDAQVSLIIFAASGKMHDYISPSTTLVDMLERY 180
Query: 71 QKCSYGAVEVSKPAKELESSYREYLKLKQRFENLQRAQRNLLGEDLGPLSSKD 123
K S + +K E+ E +LK+ + +Q R+L G+D+ L+ K+
Sbjct: 181 HKTSGKRLWDAKH----ENLNGEIERLKKENDGMQIELRHLKGDDINSLNYKE 327
>TC17763 similar to UP|Q7Y1U9 (Q7Y1U9) SVP-like floral repressor, partial
(35%)
Length = 737
Score = 80.9 bits (198), Expect = 2e-16
Identities = 39/70 (55%), Positives = 56/70 (79%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
M R ++++K+I+N RQVTF+KRR GL KKA ELSVLCDA+VAL++FS+ GKL+E+ S
Sbjct: 501 MAREKIQIKKIDNATARQVTFSKRRRGLFKKAEELSVLCDADVALVVFSSTGKLFEY-SN 677
Query: 61 SNMLKTLDRY 70
+M + L+R+
Sbjct: 678 LSMKEILERH 707
>AV421179
Length = 207
Score = 78.2 bits (191), Expect = 1e-15
Identities = 36/61 (59%), Positives = 49/61 (80%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
M R R+++K+I+N +RQVTF+KRR GL KKA ELS LCDA++ALI+FS KL+E+ S+
Sbjct: 6 MTRKRIQIKKIDNISSRQVTFSKRRKGLFKKAQELSTLCDADIALIVFSATNKLFEYASS 185
Query: 61 S 61
S
Sbjct: 186S 188
>BP064158
Length = 394
Score = 74.3 bits (181), Expect = 2e-14
Identities = 36/67 (53%), Positives = 49/67 (72%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGR +++ +++EN NRQVTF+KRRNGL+KKAYELSVLC +VALI+FS G+ F
Sbjct: 165 MGRVKLQNQKVENTTNRQVTFSKRRNGLIKKAYELSVLCXFDVALIMFSPSGRACLFSGN 344
Query: 61 SNMLKTL 67
++ L
Sbjct: 345 RRLIGEL 365
>TC11647 similar to UP|Q7X9H7 (Q7X9H7) MADS-box protein AGL62, partial (25%)
Length = 843
Score = 70.1 bits (170), Expect = 3e-13
Identities = 30/57 (52%), Positives = 45/57 (78%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEF 57
+GR ++E+K++ N+ N QVTF+KRR+GL KKA EL +LCD E+AL++FS K++ F
Sbjct: 85 LGRQKIEMKKMTNESNLQVTFSKRRSGLFKKASELCILCDVEIALVVFSPGEKVFSF 255
>AU089354
Length = 178
Score = 45.8 bits (107), Expect = 7e-06
Identities = 22/49 (44%), Positives = 35/49 (70%), Gaps = 1/49 (2%)
Frame = +3
Query: 27 GLLKKAYELSVLCDAEVALIIFSTRGKLYEFCSTS-NMLKTLDRYQKCS 74
G+LKKA E++VLC A+V+LI F+ GK++++ S ++ L+RY K S
Sbjct: 3 GILKKAIEITVLCXAQVSLIXFAASGKMHDYIRPSITXVEMLERYXKTS 149
>TC9164 weakly similar to UP|Q84MJ0 (Q84MJ0) MADS4 (Fragment), partial
(42%)
Length = 596
Score = 33.1 bits (74), Expect = 0.046
Identities = 14/49 (28%), Positives = 23/49 (46%)
Frame = +1
Query: 200 HSQSFFQQLECNPTLQIGSDYRYNNVASDQIASTSQAQQQVNGFVPGWM 248
H +F+ +EC PTLQIG D ++ + N ++ GW+
Sbjct: 88 HGDAFYHPIECEPTLQIG-------YQPDPVSVVTAGPSMNNNYMAGWL 213
>TC9326 homologue to UP|Q93WX4 (Q93WX4) Suppressor of K+ transport growth
defect-like protein (Fragment), partial (73%)
Length = 894
Score = 31.6 bits (70), Expect = 0.13
Identities = 16/60 (26%), Positives = 32/60 (52%)
Frame = -1
Query: 127 LERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEEINSRNHYRQSWEA 186
L++ + S + + S KT F ++++ +N +L + ++SLS KL ++ R W A
Sbjct: 420 LKKNIASWVLRTGSNKTSFTQTEISEPENPSVLLAKYSKSLSFKLWGVSPRCTLNMCWRA 241
>TC7928 similar to UP|Q93YR3 (Q93YR3) HSP associated protein like, partial
(52%)
Length = 765
Score = 26.6 bits (57), Expect = 4.3
Identities = 15/25 (60%), Positives = 16/25 (64%), Gaps = 2/25 (8%)
Frame = -3
Query: 2 GRGRVELKRIENKINR--QVTFAKR 24
GRGRVE RI +NR QVTF R
Sbjct: 454 GRGRVERNRICQALNRLLQVTFRNR 380
>TC11568 similar to UP|Q9AYT8 (Q9AYT8) Mangrin, partial (68%)
Length = 866
Score = 26.2 bits (56), Expect = 5.6
Identities = 23/61 (37%), Positives = 32/61 (51%), Gaps = 11/61 (18%)
Frame = -1
Query: 104 LQRAQRNLLGEDLGPLSSKDLEQLER-----QLDSSL---KQVRST---KTQFMLDQLAD 152
L R + LLG+ L L K +E LER QLD ++ K RST K +F++ Q+
Sbjct: 455 LNRLGKKLLGQVLDTLEIKCVE*LEREHNLIQLDLTIHPFKDARSTSHSKIRFLIGQVRT 276
Query: 153 L 153
L
Sbjct: 275 L 273
>AV781055
Length = 612
Score = 25.8 bits (55), Expect = 7.4
Identities = 17/55 (30%), Positives = 27/55 (48%), Gaps = 1/55 (1%)
Frame = +2
Query: 160 LVEANRSLSIKLEEINSRNHYRQSWEA-SDQSMQYEAQQNAHSQSFFQQLECNPT 213
+V ++L++ E R R S + SD + EAQQN Q+ +L NP+
Sbjct: 44 VVAMEQTLTLNPEAEAEREVMRDSANSDSDSDSEDEAQQNLQLQALETELAANPS 208
>BP077142
Length = 385
Score = 25.4 bits (54), Expect = 9.6
Identities = 11/25 (44%), Positives = 15/25 (60%), Gaps = 1/25 (4%)
Frame = -2
Query: 200 HSQSFFQQLECNPTL-QIGSDYRYN 223
HSQ F Q+ CNP + +G +YN
Sbjct: 180 HSQHVFLQINCNPLIYPLGECTKYN 106
>BP063957
Length = 479
Score = 25.4 bits (54), Expect = 9.6
Identities = 17/65 (26%), Positives = 30/65 (46%)
Frame = +1
Query: 139 RSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEEINSRNHYRQSWEASDQSMQYEAQQN 198
RST+ + + ++ H+L +RSL+ LE +S H R++ + S
Sbjct: 154 RSTRLVLEAGAHSHITSRRHLLSGIHRSLAHPLEAGSSNIHKRETVQGSTSYTTRPYVGR 333
Query: 199 AHSQS 203
AH Q+
Sbjct: 334 AHIQA 348
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.129 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,334,732
Number of Sequences: 28460
Number of extensions: 35479
Number of successful extensions: 190
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 183
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 184
length of query: 249
length of database: 4,897,600
effective HSP length: 88
effective length of query: 161
effective length of database: 2,393,120
effective search space: 385292320
effective search space used: 385292320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 54 (25.4 bits)
Medicago: description of AC137823.1


