

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137821.7 - phase: 0
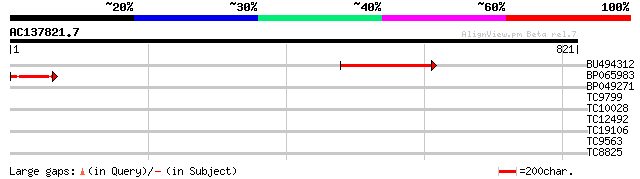
(821 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BU494312 236 1e-62
BP065983 104 5e-23
BP049271 34 0.082
TC9799 weakly similar to UP|CYB5_BRAOL (P40934) Cytochrome b5, p... 32 0.53
TC10028 weakly similar to UP|Q9LXX7 (Q9LXX7) Cytochrome P450-lik... 29 2.6
TC12492 weakly similar to UP|AAS53933 (AAS53933) AFR562Cp, parti... 29 3.4
TC19106 28 4.5
TC9563 28 7.7
TC8825 weakly similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specific ... 28 7.7
>BU494312
Length = 434
Score = 236 bits (602), Expect = 1e-62
Identities = 128/138 (92%), Positives = 132/138 (94%)
Frame = +2
Query: 480 KGATVASSQQQISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVSQYI 539
K VASSQQQISVTVVTEFVRWNEEAI+RCNLFSSQP+TLAT VKAVFT LLDQVSQYI
Sbjct: 20 KELQVASSQQQISVTVVTEFVRWNEEAISRCNLFSSQPATLATNVKAVFTSLLDQVSQYI 199
Query: 540 AEGLERARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRS 599
A+GLERARD LTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRS
Sbjct: 200 ADGLERARDSLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRS 379
Query: 600 GSSVAIIQQYFSNSISRL 617
GSSVAIIQQYF+NSISRL
Sbjct: 380 GSSVAIIQQYFANSISRL 433
>BP065983
Length = 356
Score = 104 bits (260), Expect = 5e-23
Identities = 54/69 (78%), Positives = 61/69 (88%)
Frame = -2
Query: 1 MREPRDATKTDLKTAKSASSAASFPLILDIDDFKGDFSFDALFGNLVNELLPSFKLEDLE 60
MREPRD TKTD +T+KS S ASFPLILDIDDFKGDFSFDALFGNLVNELLPSFK+E E
Sbjct: 208 MREPRDGTKTD-RTSKSTPSPASFPLILDIDDFKGDFSFDALFGNLVNELLPSFKVE-AE 35
Query: 61 AEGADAVQN 69
++GAD++ N
Sbjct: 34 SDGADSLPN 8
>BP049271
Length = 416
Score = 34.3 bits (77), Expect = 0.082
Identities = 15/17 (88%), Positives = 17/17 (99%)
Frame = -3
Query: 805 EDYKSAKLASKLSSLWS 821
EDYKSA+LASKLSSLW+
Sbjct: 414 EDYKSAQLASKLSSLWA 364
>TC9799 weakly similar to UP|CYB5_BRAOL (P40934) Cytochrome b5, partial
(64%)
Length = 799
Score = 31.6 bits (70), Expect = 0.53
Identities = 20/73 (27%), Positives = 31/73 (42%)
Frame = +3
Query: 579 EAAAAAGESSFRSFMVAVQRSGSSVAIIQQYFSNSISRLLLPVDGAHAAACEEMATAMSS 638
EA A E V S S++ +++QYF + LP G ++A ATA +
Sbjct: 201 EALITATEKDATIDFEDVGHSDSAIEMMEQYFVGEVDTNTLPSKGGSSSAAPTQATASGN 380
Query: 639 AEAAAYKGLQQCI 651
A A + Q +
Sbjct: 381 ESAGALLKILQYV 419
>TC10028 weakly similar to UP|Q9LXX7 (Q9LXX7) Cytochrome P450-like protein,
partial (16%)
Length = 874
Score = 29.3 bits (64), Expect = 2.6
Identities = 25/66 (37%), Positives = 33/66 (49%), Gaps = 4/66 (6%)
Frame = +1
Query: 14 TAKSASSAASFPLILDIDDFKGDFSFDAL----FGNLVNELLPSFKLEDLEAEGADAVQN 69
T A++AAS +LD+ D FSFD + FG LLPSF + +L AE D
Sbjct: 553 TPLMAATAASANNVLDLQDILRRFSFDNICKFSFGLDPCCLLPSFPVSEL-AEAFDLAST 729
Query: 70 KYSQVA 75
+Q A
Sbjct: 730 LSAQRA 747
>TC12492 weakly similar to UP|AAS53933 (AAS53933) AFR562Cp, partial (7%)
Length = 922
Score = 28.9 bits (63), Expect = 3.4
Identities = 28/121 (23%), Positives = 57/121 (46%), Gaps = 1/121 (0%)
Frame = +1
Query: 77 SPLFPEVEKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEK-GVDG 135
SPL +V+ + S +LLE ++Q + +H++++ V+ Q S + EL ++G
Sbjct: 508 SPLDEDVD*IDSDDNGDTSDLLEGQRQYNSAIHSIQEKVTEQPSILQG--GELRSYQIEG 681
Query: 136 LFASFARLDSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELS 195
L + ++ ++ + +G + QTI LI +LME+ G + ++
Sbjct: 682 LQWMLSLFNNNLNGILADEMGLGKTI-----------QTISLIAHLMEYKGVTGPHLIVA 828
Query: 196 P 196
P
Sbjct: 829 P 831
>TC19106
Length = 725
Score = 28.5 bits (62), Expect = 4.5
Identities = 18/42 (42%), Positives = 22/42 (51%)
Frame = -1
Query: 546 ARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGES 587
A + +T ERFV GTS S + A +AA AAA G S
Sbjct: 530 AAESMTWVLFSSERFVQGTSASMVMVLEALAAAMAAARCGLS 405
>TC9563
Length = 757
Score = 27.7 bits (60), Expect = 7.7
Identities = 16/50 (32%), Positives = 26/50 (52%), Gaps = 3/50 (6%)
Frame = +1
Query: 695 LSRVLESAFTALEGLNKQA--FLSELGNRLHKVLLN-HWQKYTFNPSGGL 741
+SR F+ L +K + + + +LH++ N W Y F+PSGGL
Sbjct: 244 ISRATSYVFSLLPASDKHFGDYAATIKKQLHEIQTN*LWGVYMFDPSGGL 393
>TC8825 weakly similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specific fungal
endoglucanase inhibitor protein precursor, partial (49%)
Length = 882
Score = 27.7 bits (60), Expect = 7.7
Identities = 24/90 (26%), Positives = 40/90 (43%), Gaps = 6/90 (6%)
Frame = +3
Query: 318 GDQAAQSSPNNVARGLSS---LYKEITDTVRKEAATITAVFPSPNEVM---SILVQRVLE 371
GD+ SP+N G+SS ++ ++ + T T V PN + S +VQ L
Sbjct: 354 GDKICGRSPSNTVTGVSSYGDIHSDVVSVNSTDGTTPTKVVSVPNFLFICGSKVVQNGLA 533
Query: 372 QRVTALLDKLLVKPSLVNLPSMEEGGLLFY 401
+ VT + + + V+LPS F+
Sbjct: 534 KGVTGMAG---LGRTRVSLPSQFSSAFSFH 614
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.129 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,175,541
Number of Sequences: 28460
Number of extensions: 109120
Number of successful extensions: 533
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 514
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 526
length of query: 821
length of database: 4,897,600
effective HSP length: 98
effective length of query: 723
effective length of database: 2,108,520
effective search space: 1524459960
effective search space used: 1524459960
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC137821.7


