

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137821.2 + phase: 0 /pseudo
(428 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
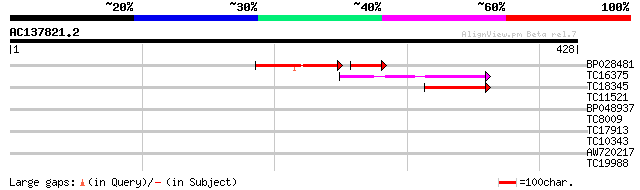
Score E
Sequences producing significant alignments: (bits) Value
BP028481 54 3e-11
TC16375 weakly similar to UP|Q9FEV9 (Q9FEV9) Microtubule-associa... 64 5e-11
TC18345 similar to UP|Q84VU1 (Q84VU1) 65 kDa microtubule associa... 40 6e-04
TC11521 similar to UP|O23144 (O23144) Proton pump interactor (AT... 32 0.15
BP048937 31 0.34
TC8009 homologue to UP|Q9LKJ2 (Q9LKJ2) Cytosolic phosphoglycerat... 28 2.2
TC17913 28 2.9
TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (... 27 4.9
AW720217 27 8.4
TC19988 similar to UP|CRT3_ARATH (O04153) Calreticulin 3 precurs... 27 8.4
>BP028481
Length = 313
Score = 53.5 bits (127), Expect(2) = 3e-11
Identities = 32/70 (45%), Positives = 49/70 (69%), Gaps = 4/70 (5%)
Frame = +1
Query: 186 ETLARLNESVLLLKQEKQQSLQKVQFLE----ELWDFMGITTDEQKAVCDDVIGLISASV 241
+TLARL ++VL LK++K+Q L K+Q L +LW+ M T E++++ D V ISASV
Sbjct: 1 DTLARLAKTVLTLKEDKKQRLHKLQELASQLIDLWNLMD-TPPEERSLFDHVTCNISASV 177
Query: 242 DEVSIHGSLS 251
DEV++ G+L+
Sbjct: 178 DEVTVPGALA 207
Score = 30.8 bits (68), Expect(2) = 3e-11
Identities = 14/27 (51%), Positives = 19/27 (69%)
Frame = +2
Query: 258 VDVEVQRLQVLNPSKMKEFVFKRQKEL 284
++VEV+RL L S+MKE K+Q EL
Sbjct: 233 LEVEVERLDQLKASRMKEIALKKQGEL 313
>TC16375 weakly similar to UP|Q9FEV9 (Q9FEV9) Microtubule-associated protein
MAP65-1a, partial (16%)
Length = 732
Score = 63.9 bits (154), Expect = 5e-11
Identities = 40/114 (35%), Positives = 64/114 (56%)
Frame = +2
Query: 250 LSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEEIHRGAHMDMDAKAAR 309
L+ D++ Q +VE++ L L S MK+ ++ K+Q +LEEI AH+++D +
Sbjct: 2 LAMDMI*QAEVEIESLGQLTTSMMKD--------ISVKKQGKLEEIFSRAHVEIDPE--- 148
Query: 310 QILTDLIGNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEWKSAAEEEKWLD 363
D I + S +L MD Q KA+ A SR+DI+D+V + A EEE W++
Sbjct: 149 ----DAIDRMGHSSILASMDSQKAKAEA*ALSRKDILDKVDK*MLAREEESWVE 298
>TC18345 similar to UP|Q84VU1 (Q84VU1) 65 kDa microtubule associated
protein, partial (8%)
Length = 521
Score = 40.4 bits (93), Expect = 6e-04
Identities = 21/50 (42%), Positives = 30/50 (60%)
Frame = +3
Query: 314 DLIGNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEWKSAAEEEKWLD 363
D I + S +L MD Q KA+ A SR+DI+D+V + A EEE W++
Sbjct: 24 DAIDRMGHSSILASMDSQKAKAEA*ALSRKDILDKVDK*MLAREEESWVE 173
>TC11521 similar to UP|O23144 (O23144) Proton pump interactor
(AT4g27500/F27G19_100), partial (21%)
Length = 1005
Score = 32.3 bits (72), Expect = 0.15
Identities = 43/208 (20%), Positives = 78/208 (36%), Gaps = 6/208 (2%)
Frame = +2
Query: 62 QLANIIPVVEDLRFKKKERVKEILEIKSQ-ISQIRAEKAGCGQSKGVTDQDVDQCDLTME 120
QL I + RF+ E +K +++ ISQI++ + Q + + D+ + + + +
Sbjct: 470 QLDKEISKINQGRFQITEALKAKRSDRAELISQIKSLRDNSRQFQSMVDEKIKEIEPLQQ 649
Query: 121 KLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNGAQ 180
LGKL++ N + L LND Q
Sbjct: 650 ALGKLRTATNAGRGG-----------------------------LCSSEEELNDVIYSFQ 742
Query: 181 QSISDETLARLNESVLL-----LKQEKQQSLQKVQFLEELWDFMGITTDEQKAVCDDVIG 235
I E+++ E +L L+ +++ + +L D MG QK D +
Sbjct: 743 YRIQHESISLAEEKQILRDIKQLEGTREKVVANAAMRAKLQDSMG-----QKDTIQDQVK 907
Query: 236 LISASVDEVSIHGSLSNDIVKQVDVEVQ 263
LI +D V +KQ+D E++
Sbjct: 908 LIGGDLDGVKKERQAIRSKIKQIDEELK 991
>BP048937
Length = 561
Score = 31.2 bits (69), Expect = 0.34
Identities = 13/32 (40%), Positives = 24/32 (74%)
Frame = -1
Query: 60 KQQLANIIPVVEDLRFKKKERVKEILEIKSQI 91
++QL +PV+E L +K+ER++E +++SQI
Sbjct: 507 EEQLETKVPVLEQLWQQKEERIQEFSDVQSQI 412
Score = 29.3 bits (64), Expect = 1.3
Identities = 16/44 (36%), Positives = 26/44 (58%)
Frame = -2
Query: 259 DVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEEIHRGAHMD 302
+VEV+RL+ S+MKE A ++Q EL+E+ AH++
Sbjct: 278 EVEVERLEQFKASRMKEI--------ASRKQGELKEMFACAHVE 171
>TC8009 homologue to UP|Q9LKJ2 (Q9LKJ2) Cytosolic phosphoglycerate kinase
, complete
Length = 1697
Score = 28.5 bits (62), Expect = 2.2
Identities = 25/90 (27%), Positives = 40/90 (43%)
Frame = -2
Query: 183 ISDETLARLNESVLLLKQEKQQSLQKVQFLEELWDFMGITTDEQKAVCDDVIGLISASVD 242
+++ + LN S L++ + + LQ V LW Q A DD+ S +D
Sbjct: 481 LTNRVASNLNLSSKKLRKPRHKWLQTVFRCYTLWP-------SQMAGKDDLSTKTSQVLD 323
Query: 243 EVSIHGSLSNDIVKQVDVEVQRLQVLNPSK 272
H S ++ I+ V V +QR +NP K
Sbjct: 322 --GRHSSTNSSIISDVKVIIQRNIQINPDK 239
>TC17913
Length = 572
Score = 28.1 bits (61), Expect = 2.9
Identities = 20/61 (32%), Positives = 32/61 (51%), Gaps = 5/61 (8%)
Frame = -3
Query: 120 EKLGKLKSHLNELQNEKNIRHQKVKSHIST-----ISELSAVMSIDISEILNGIHPSLND 174
EK L H+ +L N+KN++H+ + I T I++LS S +++ I P ND
Sbjct: 447 EKPSSLVVHVIQLHNQKNVKHKLNQGIIHTRGEERINDLSDSTSTNLT-----IKPQEND 283
Query: 175 S 175
S
Sbjct: 282 S 280
>TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (Fibrillarin
2) (AtFib2), partial (85%)
Length = 1116
Score = 27.3 bits (59), Expect = 4.9
Identities = 18/61 (29%), Positives = 28/61 (45%), Gaps = 5/61 (8%)
Frame = +1
Query: 26 YLNKVLDDFQSEIANIASSLGEDY-VSFSRGKG----TLKQQLANIIPVVEDLRFKKKER 80
YL S +++I G Y V FS G + ++ N+IP++ED R K R
Sbjct: 484 YLGAASGTNVSHVSDIVGPTGVVYAVEFSHRSGRDLVNMAKKRTNVIPIIEDARHPAKYR 663
Query: 81 V 81
+
Sbjct: 664 M 666
>AW720217
Length = 539
Score = 26.6 bits (57), Expect = 8.4
Identities = 16/52 (30%), Positives = 27/52 (51%)
Frame = +2
Query: 172 LNDSSNGAQQSISDETLARLNESVLLLKQEKQQSLQKVQFLEELWDFMGITT 223
L D S +QQ++ + L R +E+ ++Q+L E+ F+GITT
Sbjct: 341 LGDKSPFSQQNLQGDALDRDDET------------PRIQYLAEIAKFLGITT 460
>TC19988 similar to UP|CRT3_ARATH (O04153) Calreticulin 3 precursor, partial
(26%)
Length = 642
Score = 26.6 bits (57), Expect = 8.4
Identities = 11/40 (27%), Positives = 26/40 (64%)
Frame = +2
Query: 304 DAKAARQILTDLIGNIDMSELLQGMDKQIRKAKEQAQSRR 343
D AR+++ ++ N ++ + +++IRKAKE+ +++R
Sbjct: 173 DPDYAREVVDEVFANREIEKEAFEEEEKIRKAKEEEEAQR 292
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.135 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,173,783
Number of Sequences: 28460
Number of extensions: 52739
Number of successful extensions: 258
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 257
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 257
length of query: 428
length of database: 4,897,600
effective HSP length: 93
effective length of query: 335
effective length of database: 2,250,820
effective search space: 754024700
effective search space used: 754024700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC137821.2


