

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137703.11 + phase: 0
(323 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
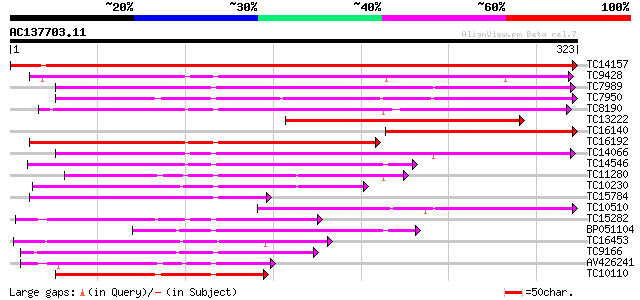
Score E
Sequences producing significant alignments: (bits) Value
TC14157 similar to UP|O24080 (O24080) Peroxidase2 precursor , p... 561 e-161
TC9428 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), pa... 234 1e-62
TC7989 similar to UP|Q43854 (Q43854) Peroxidase precursor , par... 232 7e-62
TC7950 similar to UP|Q9XFI8 (Q9XFI8) Peroxidase (Fragment), part... 222 6e-59
TC8190 similar to UP|PER1_ARAHY (P22195) Cationic peroxidase 1 p... 215 7e-57
TC13222 weakly similar to UP|Q84UA9 (Q84UA9) Peroxidase 1, parti... 179 4e-46
TC16140 similar to UP|Q40367 (Q40367) Peroxidase precursor (Frag... 175 1e-44
TC16192 similar to UP|Q9SSZ9 (Q9SSZ9) Peroxidase 1, partial (49%) 174 2e-44
TC14066 homologue to UP|O64970 (O64970) Cationic peroxidase 2 ,... 159 6e-40
TC14546 similar to UP|Q9XGV6 (Q9XGV6) Bacterial-induced peroxida... 158 1e-39
TC11280 similar to UP|Q9XFL5 (Q9XFL5) Peroxidase 4 (Fragment), p... 145 1e-35
TC10230 similar to UP|PE10_ARATH (Q9FX85) Peroxidase 10 precurso... 129 7e-31
TC15784 similar to UP|PE31_ARATH (Q9LHA7) Peroxidase 31 precurso... 119 7e-28
TC10510 similar to PIR|B56555|B56555 peroxidase , anionic, precu... 117 2e-27
TC15282 similar to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fra... 117 3e-27
BP051104 115 8e-27
TC16453 similar to UP|Q40372 (Q40372) Peroxidase precursor, part... 112 1e-25
TC9166 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, partial (49%) 111 1e-25
AV426241 104 2e-23
TC10110 weakly similar to UP|Q43854 (Q43854) Peroxidase precurso... 102 7e-23
>TC14157 similar to UP|O24080 (O24080) Peroxidase2 precursor , partial
(96%)
Length = 1390
Score = 561 bits (1447), Expect = e-161
Identities = 277/324 (85%), Positives = 300/324 (92%), Gaps = 1/324 (0%)
Frame = +1
Query: 1 MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA 60
MG+ N +LV SL+LTLC YT FAQLSPNHYA+ CPN+QSIV+ VQKKFQQTFVTVPA
Sbjct: 55 MGQINRVLVLSLSLTLCF--YTCFAQLSPNHYASTCPNLQSIVKGVVQKKFQQTFVTVPA 228
Query: 61 TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR 120
TLRLFFHDCFVQGCDASV+VASSGNNKAEKDHP+NLSLAGDGFDTVIKAKAA+DAVPQCR
Sbjct: 229 TLRLFFHDCFVQGCDASVMVASSGNNKAEKDHPDNLSLAGDGFDTVIKAKAAVDAVPQCR 408
Query: 121 NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 180
NKVSCADILALATRDV+ LAGGPSYTVELGRFDGLVSR+SDVNGRLP+P+FNLNQLN+LF
Sbjct: 409 NKVSCADILALATRDVVVLAGGPSYTVELGRFDGLVSRASDVNGRLPEPNFNLNQLNSLF 588
Query: 181 ANNGLTQTDMIALSGAHTLGFSHCDRFSNRI-QTPVDPTLNKQYAAQLQQMCPRNVDPRI 239
A+ GLTQTDMIALSGAHTLGFSHC+RFSNRI TPVDPTLN+ YA QLQQMCP+NV+P+I
Sbjct: 589 ASQGLTQTDMIALSGAHTLGFSHCNRFSNRIYSTPVDPTLNRNYATQLQQMCPKNVNPQI 768
Query: 240 AINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITA 299
AINMDPTTPRTFDN+YYKNLQQGKGLFTSDQILFTD RS+ TVNSFA+N N FNANF A
Sbjct: 769 AINMDPTTPRTFDNIYYKNLQQGKGLFTSDQILFTDQRSKATVNSFASNSNTFNANFAAA 948
Query: 300 MTKLGRVGVKNARNGKIRTDCSVL 323
M KLGRVGVK ARNGKIRTDCSVL
Sbjct: 949 MIKLGRVGVKTARNGKIRTDCSVL 1020
>TC9428 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), partial
(97%)
Length = 1354
Score = 234 bits (597), Expect = 1e-62
Identities = 138/323 (42%), Positives = 184/323 (56%), Gaps = 13/323 (4%)
Frame = +2
Query: 12 LALTLC-----LIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFF 66
LA+ C L+PY++ AQL + Y + CPNV SIVR V+ + + + +RL F
Sbjct: 56 LAVVFCVVIGALVPYSSDAQLDNSFYKDTCPNVHSIVREVVRNVSKSDPRMLGSLIRLHF 235
Query: 67 HDCFVQGCDASVLVASSGNNKAEKD-HPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSC 125
HDCFVQGCDAS+L+ + +E+ P N S+ G D V + K A++ C N VSC
Sbjct: 236 HDCFVQGCDASILLNDTSTIVSEQGAFPNNNSIRG--LDVVNQIKTAVENA--CPNTVSC 403
Query: 126 ADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGL 185
ADILALA LA G + V LGR D L + S N LP P+FNL+QLN FA L
Sbjct: 404 ADILALAAEISSVLANGVDWKVPLGRRDSLTANRSLANQNLPGPNFNLSQLNASFAVQNL 583
Query: 186 TQTDMIALSGAHTLGFSHCDRFSNRIQT-----PVDPTLNKQYAAQLQQMCPRNVDPRIA 240
TD++ALSGAHT+G C F NR+ DPTLN Y L+ +CP
Sbjct: 584 NITDLVALSGAHTIGRGQCQFFVNRLYNFSNTGNPDPTLNTTYLQTLRSICPHGGPGTTL 763
Query: 241 INMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNT--VNSFATNGNVFNANFIT 298
++DPTTP T D+ Y+ NLQ GKGLF SD +LF+ + + VNSF++N +F NF
Sbjct: 764 AHLDPTTPDTLDSQYFSNLQGGKGLFQSDPVLFSTSGAATVAIVNSFSSNPTLFFENFKA 943
Query: 299 AMTKLGRVGVKNARNGKIRTDCS 321
+M K+ R+GV G+IR C+
Sbjct: 944 SMIKMSRIGVLTGSQGEIRKPCN 1012
>TC7989 similar to UP|Q43854 (Q43854) Peroxidase precursor , partial (85%)
Length = 1352
Score = 232 bits (591), Expect = 7e-62
Identities = 120/297 (40%), Positives = 174/297 (58%), Gaps = 1/297 (0%)
Frame = +3
Query: 27 LSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSGNN 86
LS Y + CP ++SI+R ++K F + LRL FHDCFVQGCDASVL+ S +
Sbjct: 138 LSWTFYDSSCPKLESIIRKELKKVFDKDIAQAAGLLRLHFHDCFVQGCDASVLLDGSASG 317
Query: 87 KAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPSYT 146
+EKD P NL+L F + ++ ++ +C VSCADI A+A RD + L+GGP Y
Sbjct: 318 PSEKDAPPNLTLRAQAFKIIEDLRSQIEK--KCGRVVSCADITAIAARDAVFLSGGPDYE 491
Query: 147 VELGRFDGLVSRSSDVN-GRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHCD 205
+ LGR DGL + +V LP P N + A L TD+++LSG HT+G SHC
Sbjct: 492 LPLGRRDGLNFATRNVTLDNLPAPQSNTTTILNSLATKNLDPTDVVSLSGGHTIGISHCS 671
Query: 206 RFSNRIQTPVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYYKNLQQGKGL 265
F++R+ DP +++ + L+ CP + + +D +P TFDN YY +L +GL
Sbjct: 672 SFTDRLYPSKDPVMDQTFEKNLKLTCPASNTDNTTV-LDLRSPNTFDNKYYVDLMNRQGL 848
Query: 266 FTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTDCSV 322
F SDQ L+TD R+++ V SFA N ++F F+ AM K+G++ V G+IR +CSV
Sbjct: 849 FFSDQDLYTDKRTKDIVTSFAVNQSLFFEKFVVAMLKMGQLNVLTGSQGEIRANCSV 1019
>TC7950 similar to UP|Q9XFI8 (Q9XFI8) Peroxidase (Fragment), partial (86%)
Length = 1286
Score = 222 bits (566), Expect = 6e-59
Identities = 123/297 (41%), Positives = 172/297 (57%)
Frame = +1
Query: 27 LSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSGNN 86
LS + Y+ CP ++++VR+ ++K ++ P LR+FFHDCFVQGCD SVL+ S
Sbjct: 106 LSFSFYSKTCPKLETVVRNHLKKVLKKDNGQAPGLLRIFFHDCFVQGCDGSVLLDGS--- 276
Query: 87 KAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPSYT 146
E+D P N+ + + T+ +A + QC VSCADI LA+RD + L GGP Y
Sbjct: 277 PGERDQPANIGIRPEALQTIEDIRALVHK--QCGKIVSCADITILASRDAVFLTGGPDYA 450
Query: 147 VELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHCDR 206
V LGR DG VS S+ +LP P N FA+ TD++ALSGAHT G +HC
Sbjct: 451 VPLGRRDG-VSFSTVGTQKLPSPINNTTATLKAFADRNFDATDVVALSGAHTFGRAHCGT 627
Query: 207 FSNRIQTPVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYYKNLQQGKGLF 266
F NR+ +P+DP ++K A L CP A N+D TP FDN YY +L +G+F
Sbjct: 628 FFNRL-SPLDPNMDKTLAKNLTATCPAQNSTNTA-NLDIRTPNVFDNKYYLDLMNRQGVF 801
Query: 267 TSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTDCSVL 323
TSDQ L +D R++ VN+FA N +F F+ A+ KL ++ V G+IR C+V+
Sbjct: 802 TSDQDLLSDKRTKGLVNAFAVNQTLFFEKFVDAVIKLSQLDVLTGNQGEIRGRCNVV 972
>TC8190 similar to UP|PER1_ARAHY (P22195) Cationic peroxidase 1 precursor
(PNPC1) , partial (96%)
Length = 1202
Score = 215 bits (548), Expect = 7e-57
Identities = 127/306 (41%), Positives = 173/306 (56%), Gaps = 2/306 (0%)
Frame = +1
Query: 17 CLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDA 76
CLI + AQLS YA CP V + +++ V + + LRL FHDCFVQGCDA
Sbjct: 115 CLIGIGS-AQLSSTFYAKTCPLVLATIKTQVNLAVAKEARMGASLLRLHFHDCFVQGCDA 291
Query: 77 SVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDV 136
S+L+ + + EK N + G+D + K+ ++++ C VSCADI+A+A RD
Sbjct: 292 SILLDDTSSFTGEKTAGPNANSVR-GYDVIDTIKSKVESL--CPGVVSCADIVAVAARDS 462
Query: 137 INLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGA 196
+ GG S+ V LGR D + S N LP PS NL+ LNT F+N G T +M+ALSG+
Sbjct: 463 VVALGGFSWAVPLGRRDSTTASLSSANSELPGPSSNLDGLNTAFSNKGFTTREMVALSGS 642
Query: 197 HTLGFSHCDRFSNRI--QTPVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNV 254
HT+G + C F RI +T +D T +A LQ CP N +D T+P TFDN
Sbjct: 643 HTIGQARCLFFRTRIYYETHIDST----FAKNLQGNCPFNGGDSNLSPLDTTSPTTFDNG 810
Query: 255 YYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNG 314
YY+NLQ KGLF SDQ+LF + + VNS+ TN F +F AM K+G + +G
Sbjct: 811 YYRNLQSQKGLFHSDQVLFNGGSTDSQVNSYVTNPASFKTDFANAMVKMGNLSPLTGSSG 990
Query: 315 KIRTDC 320
+IRT C
Sbjct: 991 QIRTHC 1008
>TC13222 weakly similar to UP|Q84UA9 (Q84UA9) Peroxidase 1, partial (41%)
Length = 421
Score = 179 bits (455), Expect = 4e-46
Identities = 87/137 (63%), Positives = 104/137 (75%), Gaps = 1/137 (0%)
Frame = +2
Query: 158 RSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHCDRFSNRI-QTPVD 216
++S V G LP+P+FNLNQLNT+F+N+ LTQTDMIALSGAHT+GFSHC+ FSNRI +PVD
Sbjct: 5 KASRVAGNLPKPNFNLNQLNTMFSNHNLTQTDMIALSGAHTVGFSHCNEFSNRIYSSPVD 184
Query: 217 PTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDT 276
PTL+ Y+ QL CP+N DP + + +DP T TFDN YYKNL GKGL SDQ+LFTD
Sbjct: 185 PTLDPTYSQQLIAECPKNPDPGVVVALDPETSATFDNEYYKNLVAGKGLLASDQVLFTDP 364
Query: 277 RSRNTVNSFATNGNVFN 293
SR TV FA NG FN
Sbjct: 365 ASRATVVEFANNGGEFN 415
>TC16140 similar to UP|Q40367 (Q40367) Peroxidase precursor (Fragment) ,
partial (34%)
Length = 484
Score = 175 bits (443), Expect = 1e-44
Identities = 79/109 (72%), Positives = 95/109 (86%)
Frame = +3
Query: 215 VDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFT 274
VDPT+N+QYA +L+ CPRNVDPRIAINMDPTTP+ FDNVY++NLQ+GKGLFTSDQ+LF
Sbjct: 9 VDPTMNRQYATELKSPCPRNVDPRIAINMDPTTPKAFDNVYFQNLQKGKGLFTSDQVLFM 188
Query: 275 DTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTDCSVL 323
D+RS+ VN+FA++ F ANF+ AMTKLGRVGVKNA NG IRTDCSV+
Sbjct: 189 DSRSKAAVNAFASSSKTFRANFVAAMTKLGRVGVKNAHNGNIRTDCSVI 335
>TC16192 similar to UP|Q9SSZ9 (Q9SSZ9) Peroxidase 1, partial (49%)
Length = 645
Score = 174 bits (440), Expect = 2e-44
Identities = 90/200 (45%), Positives = 125/200 (62%)
Frame = +2
Query: 12 LALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFV 71
+ L + L+ +++L +Y+ C + IV+ V++ + +R+ FHDCF+
Sbjct: 38 IVLVIFLLNQNAYSELEVGYYSYSCGMAEFIVKDEVRRSVTKNPGIAAGLVRMHFHDCFI 217
Query: 72 QGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILAL 131
+GCDASVL+ S+ +N AEKD P N GF+ + AKA L+AV C+ VSCADI+A
Sbjct: 218 RGCDASVLLDSTPSNTAEKDSPANKPSLR-GFEVIDSAKAKLEAV--CKGVVSCADIIAF 388
Query: 132 ATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMI 191
A RD + LAGG Y V GR DG +S +SD LP P+FN+NQL LFA GLTQ +M+
Sbjct: 389 AARDSVELAGGVGYDVPAGRRDGRISLASDTRTDLPPPTFNVNQLTQLFAKKGLTQEEMV 568
Query: 192 ALSGAHTLGFSHCDRFSNRI 211
LSGAHT+G SHC FS+R+
Sbjct: 569 TLSGAHTIGRSHCAAFSSRL 628
>TC14066 homologue to UP|O64970 (O64970) Cationic peroxidase 2 , partial
(92%)
Length = 1436
Score = 159 bits (402), Expect = 6e-40
Identities = 99/299 (33%), Positives = 149/299 (49%), Gaps = 3/299 (1%)
Frame = +2
Query: 27 LSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSGNN 86
L+ N Y CP + I+ V+ +++ T + LR FHDC VQ CDAS+L+ S+
Sbjct: 152 LAMNFYKETCPQAEDIITEQVKLLYKRHKNTAFSWLRNIFHDCAVQSCDASLLLDSTRKT 331
Query: 87 KAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPSYT 146
+EK+ + L F + K AL+ +C VSC+DIL L+ RD I GGP
Sbjct: 332 LSEKETDRSFGLRN--FRYIETIKEALER--ECPGVVSCSDILVLSARDGIAALGGPHIP 499
Query: 147 VELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHCDR 206
+ GR DG SR+ V LP + +++ + F G+ ++AL GAH++G +HC +
Sbjct: 500 LRTGRRDGRRSRADVVEQFLPDHNESISAVLDKFGAMGIDTPGVVALLGAHSVGRTHCVK 679
Query: 207 FSNRIQTPVDPTLNKQYAAQLQQMCPRNV-DPRIA--INMDPTTPRTFDNVYYKNLQQGK 263
+R+ VDP LN + + + CP + DP+ + D TP DN YY+N+ K
Sbjct: 680 LVHRLYPEVDPALNPDHIPHMLKKCPDAIPDPKAVQYVRNDRGTPMILDNNYYRNILDNK 859
Query: 264 GLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTDCSV 322
GL D L D R++ V A + + F F A+T L G+IR C+V
Sbjct: 860 GLLLVDHQLANDKRTKPYVKKMAKSQDYFFKEFSRAITLLSENNPLTGTKGEIRKQCNV 1036
>TC14546 similar to UP|Q9XGV6 (Q9XGV6) Bacterial-induced peroxidase
precursor , partial (66%)
Length = 738
Score = 158 bits (400), Expect = 1e-39
Identities = 89/223 (39%), Positives = 131/223 (57%), Gaps = 1/223 (0%)
Frame = +1
Query: 11 SLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCF 70
+L++ L L+ + AQL+P Y CP +Q+IVR+A+ + ++ + LRLFFHDCF
Sbjct: 52 ALSILLSLLACSANAQLTPTFYDRSCPKLQTIVRNAMVQTIKKEARMGASILRLFFHDCF 231
Query: 71 VQGCDASVLVASSGNN-KAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADIL 129
V GCD S+L+ G EK+ N + A GF+ + K ++A C N VSCADIL
Sbjct: 232 VNGCDGSILLDDIGTTFVGEKNAAPNKNSAR-GFEVIDTIKTNVEA--SCNNTVSCADIL 402
Query: 130 ALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTD 189
ALATRD INL GGP++ V LGR D + S N +P PS +L+ L ++F+ GL+ D
Sbjct: 403 ALATRDGINLLGGPTWQVPLGRRDARTASQSKANTEIPSPSSDLSTLISMFSAKGLSARD 582
Query: 190 MIALSGAHTLGFSHCDRFSNRIQTPVDPTLNKQYAAQLQQMCP 232
+ LSG HT+G + C F +R+ + ++ +AA + CP
Sbjct: 583 LTVLSGGHTIGQAECQFFRSRVNN--ETNIDAAFAASRKTNCP 705
>TC11280 similar to UP|Q9XFL5 (Q9XFL5) Peroxidase 4 (Fragment), partial
(68%)
Length = 614
Score = 145 bits (365), Expect = 1e-35
Identities = 87/200 (43%), Positives = 114/200 (56%), Gaps = 4/200 (2%)
Frame = +3
Query: 32 YANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSGNNKAEKD 91
Y CP +SIVRS V+ LR+ FHDCFVQGCDASVL+A +G E+
Sbjct: 39 YLGTCPRAESIVRSTVESHVNSDPTLAAGLLRMHFHDCFVQGCDASVLIAGAG---TERT 209
Query: 92 HPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPSYTVELGR 151
NLSL GF+ + AKA ++A C VSCADILALA RD + L+GG S+ V GR
Sbjct: 210 AIPNLSLR--GFEVIDDAKAKVEAA--CPGVVSCADILALAARDSVVLSGGLSWQVPTGR 377
Query: 152 FDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHCDRFSNRI 211
DG VS++SDVN LP P +++ F GL D++ L G HT+G + C FSNR+
Sbjct: 378 RDGRVSQASDVN-NLPAPFDSVDVQKQKFTAKGLNTQDLVTLVGGHTIGTTACQFFSNRL 554
Query: 212 ----QTPVDPTLNKQYAAQL 227
DP+++ + QL
Sbjct: 555 YNFTSNGPDPSIDASFLLQL 614
>TC10230 similar to UP|PE10_ARATH (Q9FX85) Peroxidase 10 precursor (Atperox
P10) (ATP5a) , partial (42%)
Length = 753
Score = 129 bits (324), Expect = 7e-31
Identities = 76/191 (39%), Positives = 103/191 (53%)
Frame = +3
Query: 14 LTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQG 73
L L L+ +QL N Y CPN+ IVR + + LRL FHDCFV G
Sbjct: 186 LWLVLLSPLVSSQLYYNFYDTTCPNLTRIVRYNLLSAMSNDTRIAASLLRLHFHDCFVNG 365
Query: 74 CDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALAT 133
CD SVL+ + K EK+ N + + GF+ + K+AL+ C + VSCADIL LA
Sbjct: 366 CDGSVLLDDTSTQKGEKNALPNKN-SIRGFEVIDTIKSALEKA--CPSTVSCADILTLAA 536
Query: 134 RDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIAL 193
R+ + L+ GP ++V LGR DG + S+ N LP P L + F + GL + D+ L
Sbjct: 537 REAVYLSRGPFWSVPLGRRDGTTASESEAN-NLPSPFEPLENITAKFISKGLEKKDVAVL 713
Query: 194 SGAHTLGFSHC 204
SGAHT GF+ C
Sbjct: 714 SGAHTFGFAQC 746
>TC15784 similar to UP|PE31_ARATH (Q9LHA7) Peroxidase 31 precursor (Atperox
P31) (ATP41) , partial (38%)
Length = 475
Score = 119 bits (298), Expect = 7e-28
Identities = 66/139 (47%), Positives = 84/139 (59%), Gaps = 1/139 (0%)
Frame = +3
Query: 12 LALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCF- 70
L TL + A+L+ + Y + CP I+R V K T T PATLRLF HDC
Sbjct: 63 LLSTLSFFTTLSHARLTLDFYKHTCPQFSQIIRDTVTNKQITTPTTGPATLRLFLHDCLH 242
Query: 71 VQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILA 130
GCDAS+L++S+ NKAE+D NLSL GD FD V++AK AL+ C N VSCADIL
Sbjct: 243 PNGCDASILLSSTSFNKAERDADINLSLPGDAFDLVVRAKTALELA--CPNTVSCADILT 416
Query: 131 LATRDVINLAGGPSYTVEL 149
AT D++ + GGP + V L
Sbjct: 417 AATSDLLTMLGGPHFPVFL 473
>TC10510 similar to PIR|B56555|B56555 peroxidase , anionic, precursor - wood
tobacco {Nicotiana sylvestris;} , partial (53%)
Length = 764
Score = 117 bits (294), Expect = 2e-27
Identities = 69/186 (37%), Positives = 100/186 (53%), Gaps = 4/186 (2%)
Frame = +3
Query: 142 GPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGF 201
GPS+TV+LGR D + S N LP + +L L + F N GLT DM+ LSGAHT+G
Sbjct: 3 GPSWTVKLGRRDSTTASRSLANTELPSFTDDLQTLISRFDNKGLTARDMVILSGAHTIGQ 182
Query: 202 SHCDRFSNRIQTPVDPTLNKQYAAQLQQMCPRNV----DPRIAINMDPTTPRTFDNVYYK 257
+ C F RI ++ + + ++ CP + D ++A +D TP +FDN Y+K
Sbjct: 183 AQCSTFRGRIYNNASD-IDAGFGSTRRRGCPSTISVENDKKLAA-LDLVTPNSFDNNYFK 356
Query: 258 NLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIR 317
NL Q KGL SDQ+LF + V+ ++ N F ++F AM K+G + G IR
Sbjct: 357 NLIQKKGLLRSDQVLFRGGSTDTIVSEYSKNPTTFKSDFAAAMIKMGDIQPLTGSAGIIR 536
Query: 318 TDCSVL 323
CS +
Sbjct: 537 KICSAI 554
>TC15282 similar to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fragment) ,
partial (47%)
Length = 604
Score = 117 bits (292), Expect = 3e-27
Identities = 67/175 (38%), Positives = 98/175 (55%)
Frame = +1
Query: 4 YNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLR 63
+ V++V LAL + AQL Y CP + I+ V + A +R
Sbjct: 109 FKVLIVCLLALIA-----SNHAQLEQGFYTKTCPKAEKIILDFVHEHIHNAPSLAAALIR 273
Query: 64 LFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKV 123
+ FHDCFV+GCDAS+L+ S+ + +AE+D P NL++ G FD + + K+ ++A QC V
Sbjct: 274 MNFHDCFVRGCDASILLNST-SKQAERDAPPNLTVRG--FDFIDRIKSLVEA--QCPGVV 438
Query: 124 SCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNT 178
SCADI+ALA RD I GGP + V GR DG++S + ++P P+ N L T
Sbjct: 439 SCADIIALAARDSIVATGGPFWKVPTGRRDGVISNLVEARSQIPAPTSNFTTLQT 603
>BP051104
Length = 566
Score = 115 bits (289), Expect = 8e-27
Identities = 68/164 (41%), Positives = 92/164 (55%)
Frame = +2
Query: 71 VQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILA 130
V GCDAS+L + N E+ N + + GF+ + KA L+ QC VSCAD+LA
Sbjct: 71 VNGCDASILXDDTSNFIGEQTAAAN-NRSARGFNVIDGIKANLEK--QCPGVVSCADVLA 241
Query: 131 LATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDM 190
LA RD + GGPS+ V LGR D + N +P P +L+ L T FAN GL+ TD+
Sbjct: 242 LAARDSVVQLGGPSWEVGLGRRDSTTASRGTANNTIPGPFLSLSGLITNFANQGLSVTDL 421
Query: 191 IALSGAHTLGFSHCDRFSNRIQTPVDPTLNKQYAAQLQQMCPRN 234
+ALSGAHT+G + C F I D ++ YA L+ CPR+
Sbjct: 422 VALSGAHTIGLAQCKNFRAHIYN--DSNIDASYAKFLKX*CPRS 547
>TC16453 similar to UP|Q40372 (Q40372) Peroxidase precursor, partial (49%)
Length = 581
Score = 112 bits (279), Expect = 1e-25
Identities = 71/184 (38%), Positives = 99/184 (53%), Gaps = 2/184 (1%)
Frame = +2
Query: 3 RYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATL 62
R + +++ + L LIP +T A L+P +Y +CP I++S VQK + + L
Sbjct: 38 RQSFLVLVLVTLATFLIP-STLAGLTPGYYDKVCPQALPIIKSVVQKAINREQRIGASLL 214
Query: 63 RLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNK 122
RL FHDCFV GCD S+L+ + + EK NL+ + GF+ V + KAA+D + R
Sbjct: 215 RLHFHDCFVNGCDGSLLLDDTSSFVGEKTALPNLN-SVRGFEVVDEIKAAVDRACK-RPV 388
Query: 123 VSCADILALATRDVINLAGGPS--YTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 180
VSCADILA+A RD + + GG Y V LGR D + + N LP P N QL F
Sbjct: 389 VSCADILAIAARDSVAILGGKQYWYQVRLGRRDARSASQAAANSNLPPPFLNFTQLLPAF 568
Query: 181 ANNG 184
G
Sbjct: 569 QAQG 580
>TC9166 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, partial (49%)
Length = 559
Score = 111 bits (278), Expect = 1e-25
Identities = 71/171 (41%), Positives = 98/171 (56%), Gaps = 1/171 (0%)
Frame = +3
Query: 7 ILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFF 66
I V SL + L LI + AQLS N Y CP+V + V+S V + + LRLFF
Sbjct: 54 IFVLSLFM-LFLIGSSNSAQLSENFYVKKCPSVFNAVKSVVHSAVAKEARMGGSLLRLFF 230
Query: 67 HDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCA 126
HDCFV GCD SVL+ + + K EK P N S + GFD + K+ ++AV C VSCA
Sbjct: 231 HDCFVNGCDGSVLLDDTSSFKGEKTAPPN-SNSLRGFDVIDAIKSKVEAV--CPGVVSCA 401
Query: 127 DILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVN-GRLPQPSFNLNQL 176
D++A+A RD + + GGP + V+LGR D + + N G +P P +L+ L
Sbjct: 402 DVVAIAARDSVAILGGPYWKVKLGRRDSKTASFNAANSGVIPSPFSSLSDL 554
>AV426241
Length = 429
Score = 104 bits (259), Expect = 2e-23
Identities = 66/147 (44%), Positives = 87/147 (58%), Gaps = 2/147 (1%)
Frame = +3
Query: 7 ILVWSLALTLCLIPYTTFAQ--LSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRL 64
+LV+ + LTL FA S Y+ CP+++SIV+S V + F LRL
Sbjct: 21 LLVFLIVLTL-----QAFAVHGTSVGFYSKSCPSIESIVKSTVASHVKTDFEYAAGLLRL 185
Query: 65 FFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVS 124
F DCFV+GCDAS+L+A +G EK P N SL G++ + +AKA L+A QC VS
Sbjct: 186 HFRDCFVRGCDASILIAGNG---TEKQAPPNRSL--KGYEVIDEAKAKLEA--QCPGVVS 344
Query: 125 CADILALATRDVINLAGGPSYTVELGR 151
CADILALA RD + L+GG S+ V GR
Sbjct: 345 CADILALAARDSVVLSGGLSWQVPTGR 425
>TC10110 weakly similar to UP|Q43854 (Q43854) Peroxidase precursor ,
partial (34%)
Length = 497
Score = 102 bits (255), Expect = 7e-23
Identities = 51/121 (42%), Positives = 77/121 (63%)
Frame = +1
Query: 27 LSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSGNN 86
LS N YA+ CP ++ IVR+ ++K F+Q P LR+FFHDCFVQGCD S+L+ S
Sbjct: 145 LSWNFYASSCPVLEVIVRNHLKKVFKQDSGQAPGLLRIFFHDCFVQGCDGSILLDGS--- 315
Query: 87 KAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPSYT 146
+E+D P N+ + + T+ ++ + QC N VSCAD++ LA R+ ++L+GGP +
Sbjct: 316 PSERDQPANVGIRPEALKTIEVLRSLVQR--QCGNVVSCADLVVLAAREAVSLSGGPKFD 489
Query: 147 V 147
V
Sbjct: 490 V 492
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,053,643
Number of Sequences: 28460
Number of extensions: 60842
Number of successful extensions: 408
Number of sequences better than 10.0: 98
Number of HSP's better than 10.0 without gapping: 353
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 356
length of query: 323
length of database: 4,897,600
effective HSP length: 90
effective length of query: 233
effective length of database: 2,336,200
effective search space: 544334600
effective search space used: 544334600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC137703.11


