

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137603.6 - phase: 0 /pseudo
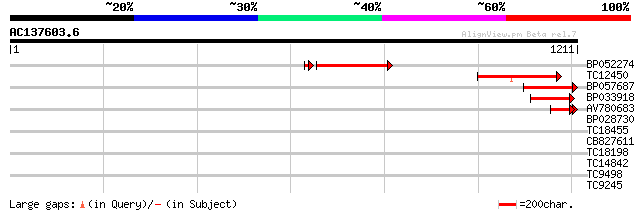
(1211 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP052274 317 1e-93
TC12450 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like prot... 276 1e-74
BP057687 187 9e-48
BP033918 140 2e-33
AV780683 88 3e-18
BP028730 33 0.27
TC18455 32 0.78
CB827611 31 1.0
TC18198 weakly similar to UP|Q9XGZ5 (Q9XGZ5) T1N24.20 protein, p... 31 1.3
TC14842 similar to UP|RK4_TOBAC (O80361) 50S ribosomal protein L... 30 3.0
TC9498 28 8.6
TC9245 28 8.6
>BP052274
Length = 563
Score = 317 bits (812), Expect(2) = 1e-93
Identities = 156/162 (96%), Positives = 160/162 (98%)
Frame = +3
Query: 656 AWFMSNQETSFVTIGQRLLANPLRVRFHYGHPDVFDRIFHLTRGGVSKASKVINLSEDIF 715
AWFMSNQETSFVTIGQRLLANPL+VRFHYGHPDVFDR+FHLTRGGVSKA +VINLSEDIF
Sbjct: 81 AWFMSNQETSFVTIGQRLLANPLKVRFHYGHPDVFDRLFHLTRGGVSKAWEVINLSEDIF 260
Query: 716 AGFNSTLREGNVTHHEYIQVGKGRDVGLNQISMFEAKIANGNGEQTLSRDVYRLGHRFDF 775
AGFNSTLREGNVTHHEYIQVGKGRDVGLNQISMFEAKIANGNGEQTLSRDVYRLGHRFDF
Sbjct: 261 AGFNSTLREGNVTHHEYIQVGKGRDVGLNQISMFEAKIANGNGEQTLSRDVYRLGHRFDF 440
Query: 776 FRMLSCYFTTIGFYFSTLQITVLTVYVFLYGRLYLVLSGLEE 817
FRMLSCYFTT+GFYFSTL ITVLTVYVFLYGRLYLVLSGLEE
Sbjct: 441 FRMLSCYFTTVGFYFSTL-ITVLTVYVFLYGRLYLVLSGLEE 563
Score = 44.7 bits (104), Expect(2) = 1e-93
Identities = 19/20 (95%), Positives = 20/20 (100%)
Frame = +1
Query: 630 KKHDGVRYPSILGLREHIFT 649
KKHDGVR+PSILGLREHIFT
Sbjct: 1 KKHDGVRFPSILGLREHIFT 60
>TC12450 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like protein
(Fragment), partial (20%)
Length = 570
Score = 276 bits (706), Expect = 1e-74
Identities = 139/188 (73%), Positives = 155/188 (81%), Gaps = 10/188 (5%)
Frame = +1
Query: 1000 GGIGVPPEKSWESWWEEEQEHLKYSGMRGIIAEILLSLRFFIYQYGLVYHLNFTKS-TKS 1058
G GVPPEKSWESWWEEEQ+HL YSG+RGII EILLSLRFFIYQYGLVYHL FTK KS
Sbjct: 7 GTRGVPPEKSWESWWEEEQDHLHYSGLRGIILEILLSLRFFIYQYGLVYHLKFTKKGDKS 186
Query: 1059 VLPTGLWHIMVG---------DLSNISYLEDFQLVFRLMKGLVFVTFVSILVTMIALAHM 1109
L G+ +++ + + +FQLVFRL+KG++F+TFVSILVT+IAL HM
Sbjct: 187 FLVYGISWLVIFVILFVMXTVSVGRRKFSANFQLVFRLIKGMIFLTFVSILVTLIALPHM 366
Query: 1110 TLQDIVVCILAFMPTGWGMLQIAQALKPLVRRGGFWESVKTLARGYEVIMGLLLFTPVAF 1169
TLQDIVVCILAFMPTGWGMLQIAQALKPLVRR GFW SVKTLARGYE++MGLLLFTPVAF
Sbjct: 367 TLQDIVVCILAFMPTGWGMLQIAQALKPLVRRAGFWGSVKTLARGYEIVMGLLLFTPVAF 546
Query: 1170 LAWFPFVS 1177
LAWFPFVS
Sbjct: 547 LAWFPFVS 570
>BP057687
Length = 573
Score = 187 bits (475), Expect = 9e-48
Identities = 86/114 (75%), Positives = 104/114 (90%)
Frame = -2
Query: 1098 SILVTMIALAHMTLQDIVVCILAFMPTGWGMLQIAQALKPLVRRGGFWESVKTLARGYEV 1157
+I + +IA+A MT++DI+VCILA MPTGWG+L IAQA KP++++GGFW SV+ LARGYE+
Sbjct: 572 TIFIVLIAVAKMTIKDIIVCILAVMPTGWGLLLIAQACKPVIKKGGFWGSVRALARGYEI 393
Query: 1158 IMGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISRILGGQRKGRSSRNK 1211
IMGLLLFTP+AFLAWFPFVSEFQTRMLFNQAFSRGLQISRILGGQ++ R+S NK
Sbjct: 392 IMGLLLFTPIAFLAWFPFVSEFQTRMLFNQAFSRGLQISRILGGQKRDRASNNK 231
>BP033918
Length = 512
Score = 140 bits (352), Expect = 2e-33
Identities = 65/94 (69%), Positives = 78/94 (82%)
Frame = -3
Query: 1113 DIVVCILAFMPTGWGMLQIAQALKPLVRRGGFWESVKTLARGYEVIMGLLLFTPVAFLAW 1172
DI V +LAF+PT W ++QIAQA +PLV+ G W SVK LARGYE +MG+++FTPVA LAW
Sbjct: 507 DIFVSLLAFLPTAWALIQIAQACRPLVKGVGMWGSVKALARGYEYLMGVVIFTPVAILAW 328
Query: 1173 FPFVSEFQTRMLFNQAFSRGLQISRILGGQRKGR 1206
FPFVSEFQTR+LFNQAFSRGLQI RIL G +K +
Sbjct: 327 FPFVSEFQTRLLFNQAFSRGLQIQRILSGGKKNK 226
>AV780683
Length = 482
Score = 88.2 bits (217), Expect(2) = 3e-18
Identities = 42/50 (84%), Positives = 46/50 (92%)
Frame = -3
Query: 1156 EVIMGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISRILGGQRKG 1205
E +M LLLFTPVAFLAWFPF+SEFQTRMLFNQAFSRGLQISRILG ++G
Sbjct: 480 EFVMXLLLFTPVAFLAWFPFLSEFQTRMLFNQAFSRGLQISRILGRPKEG 331
Score = 21.6 bits (44), Expect(2) = 3e-18
Identities = 10/13 (76%), Positives = 10/13 (76%)
Frame = -1
Query: 1199 LGGQRKGRSSRNK 1211
L GQRKGRSS K
Sbjct: 350 LDGQRKGRSSLYK 312
>BP028730
Length = 454
Score = 33.1 bits (74), Expect = 0.27
Identities = 19/64 (29%), Positives = 33/64 (50%), Gaps = 6/64 (9%)
Frame = -3
Query: 972 PFLFNPSGFE-WQKIVDDWTDWNKWISIRGGIG---VPPEKSWESWWEEEQ--EHLKYSG 1025
PF+F+ G + W D +D ++W + GG+G V S++ W+EE + L++
Sbjct: 338 PFVFDAHGIDAWDDDCSDVSDASEWDFVDGGMGEYYVDDSDSYDGMWDEEGRIDELQFGF 159
Query: 1026 MRGI 1029
GI
Sbjct: 158 YEGI 147
>TC18455
Length = 565
Score = 31.6 bits (70), Expect = 0.78
Identities = 19/55 (34%), Positives = 28/55 (50%), Gaps = 2/55 (3%)
Frame = -1
Query: 358 TPYYT-EEVLFSLRELESPNEDGVSILFYLQKIF-PDEWNNFLQRVNCSNEEELK 410
TPY ++ L L SPN +L++ F PD+W LQR NC+ + +K
Sbjct: 394 TPYVGWSTIVAKLG*LRSPNY----LLYFTSLSF*PDQWKKKLQRTNCTEQNRIK 242
>CB827611
Length = 559
Score = 31.2 bits (69), Expect = 1.0
Identities = 18/62 (29%), Positives = 29/62 (46%)
Frame = +3
Query: 788 FYFSTLQITVLTVYVFLYGRLYLVLSGLEEGLSTQKAIRDNKPLQVALASQSFVQIGFLM 847
F+ T+L +YVF + Y+ + + KAIR P V L + SF+ + F+
Sbjct: 228 FFLFVSSATILCIYVFSFSAFYIKVLMDNNDSTVWKAIR-KSPASVILMAYSFISLWFVG 404
Query: 848 AL 849
L
Sbjct: 405 GL 410
>TC18198 weakly similar to UP|Q9XGZ5 (Q9XGZ5) T1N24.20 protein, partial
(14%)
Length = 592
Score = 30.8 bits (68), Expect = 1.3
Identities = 25/101 (24%), Positives = 50/101 (48%), Gaps = 5/101 (4%)
Frame = -3
Query: 301 WTEKIKRLYLLLTTKESAMDVPSNLEAKRRISFFSNSLFMDMPTAPKVRNMLSFSVLTPY 360
W E K++ E+A++ S++ + + + L P V + L+ +
Sbjct: 479 WVEGQKKV------NEAAVNYFSDIYSSSPLQYLRECL---APIPKLVTDRLNHKLCAQV 327
Query: 361 YTEEV---LFSLRELESPNEDGVSILFYLQ--KIFPDEWNN 396
T+E+ +FS+ +L++P DG++ LFY Q +I ++ NN
Sbjct: 326 STDEIKEAVFSMGDLKAPGMDGINGLFYQQNREIVKNDVNN 204
>TC14842 similar to UP|RK4_TOBAC (O80361) 50S ribosomal protein L4,
chloroplast precursor (R-protein L4), partial (51%)
Length = 618
Score = 29.6 bits (65), Expect = 3.0
Identities = 15/39 (38%), Positives = 21/39 (53%)
Frame = +3
Query: 322 PSNLEAKRRISFFSNSLFMDMPTAPKVRNMLSFSVLTPY 360
P +SFFS+S+F+ P+ KV LS S TP+
Sbjct: 72 PRMASTSTSLSFFSSSIFLSNPSKLKVPTKLSNSNSTPF 188
>TC9498
Length = 466
Score = 28.1 bits (61), Expect = 8.6
Identities = 12/35 (34%), Positives = 19/35 (54%)
Frame = -3
Query: 388 KIFPDEWNNFLQRVNCSNEEELKEYDELEEELRRW 422
K+FP WN R++CS ++ + +E E RW
Sbjct: 368 KLFPCFWNRAWCRLHCSKQQRKQHREEQE*GSGRW 264
>TC9245
Length = 583
Score = 28.1 bits (61), Expect = 8.6
Identities = 21/66 (31%), Positives = 35/66 (52%), Gaps = 11/66 (16%)
Frame = -2
Query: 1108 HMTLQDIVVCILAFMPTGWGMLQI--------AQALK---PLVRRGGFWESVKTLARGYE 1156
H+ +DI++ ++ +P WG LQ+ Q LK PLV GF++S KTL +
Sbjct: 294 HVIPKDIIMVVIEVVPLCWGFLQLILKNFPIPCQKLKKSRPLV--DGFFQS-KTLGSTKQ 124
Query: 1157 VIMGLL 1162
++ +L
Sbjct: 123 ILEDIL 106
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.324 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,938,439
Number of Sequences: 28460
Number of extensions: 277531
Number of successful extensions: 1299
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1281
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1296
length of query: 1211
length of database: 4,897,600
effective HSP length: 101
effective length of query: 1110
effective length of database: 2,023,140
effective search space: 2245685400
effective search space used: 2245685400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC137603.6


