

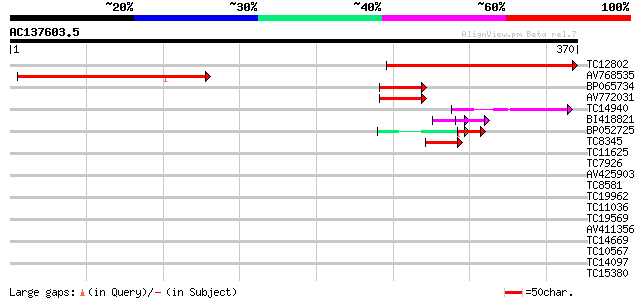
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137603.5 + phase: 0
(370 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC12802 245 1e-65
AV768535 196 7e-51
BP065734 56 8e-09
AV772031 48 2e-06
TC14940 42 2e-04
BI418821 30 5e-04
BP052725 31 7e-04
TC8345 40 8e-04
TC11625 weakly similar to UP|O81015 (O81015) At2g26920 protein, ... 36 0.009
TC7926 homologue to UP|PGKH_TOBAC (Q42961) Phosphoglycerate kina... 36 0.012
AV425903 35 0.026
TC8581 similar to UP|Q94II4 (Q94II4) NAM-like protein, partial (9%) 34 0.034
TC19962 27 0.042
TC11036 similar to UP|COQ4_ARATH (Q9ZPR0) Ubiquinone biosynthesi... 34 0.044
TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor k... 29 0.068
AV411356 33 0.076
TC14669 similar to PIR|E86326|E86326 protein F18O14.3 [imported]... 33 0.099
TC10567 similar to UP|AAR99376 (AAR99376) Ring domain containing... 33 0.099
TC14097 homologue to UP|GCSP_PEA (P26969) Glycine dehydrogenase ... 27 0.12
TC15380 similar to UP|Q8H5P5 (Q8H5P5) OJ1003_C06.27 protein, par... 32 0.13
>TC12802
Length = 574
Score = 245 bits (625), Expect = 1e-65
Identities = 109/124 (87%), Positives = 120/124 (95%)
Frame = +1
Query: 247 GALLSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSAEERMSAPTVYPPPPPPPPPKVAKEG 306
GALLSC+YTVLDYAQTGLIAAVFFFKMMEWWYQSAEERMSAPTVYPPPPPPPPPKVAKEG
Sbjct: 1 GALLSCSYTVLDYAQTGLIAAVFFFKMMEWWYQSAEERMSAPTVYPPPPPPPPPKVAKEG 180
Query: 307 VQLPSDRTICPLCLQKRVNPSVMTVSGFVFCYACIFKFLTQYKRCPATMVPATVDQIRRL 366
V LP DR +CPLCLQKRVNPSV++VSGFVFCYAC+FK+++QYKRCP T++P+TVDQIRRL
Sbjct: 181 VPLPPDRAVCPLCLQKRVNPSVISVSGFVFCYACVFKYISQYKRCPVTLMPSTVDQIRRL 360
Query: 367 FHDV 370
FHDV
Sbjct: 361 FHDV 372
>AV768535
Length = 393
Score = 196 bits (497), Expect = 7e-51
Identities = 99/130 (76%), Positives = 117/130 (89%), Gaps = 4/130 (3%)
Frame = +3
Query: 6 GGQGSRPTFFEMAAAEQLPRSLRAALTYSIGVLALRTPFLHKLLDYEHESFSLLMLVLEA 65
GGQGSRPTFFEMAAAEQLP SLRAALTYSIGVLALR PFLH+ LDYE ESF+ LML+LE+
Sbjct: 3 GGQGSRPTFFEMAAAEQLPASLRAALTYSIGVLALRRPFLHRFLDYEDESFAFLMLLLES 182
Query: 66 HSLRTTDASFSESLYGLRRRPANIKLNDNDSSSSS----SQLRRRQKLLSLLFLVVLPYL 121
HSLRTTDASFSESLYGLRRRPAN+ ++ +D+++++ S L RRQK+LS++FLVVLPYL
Sbjct: 183 HSLRTTDASFSESLYGLRRRPANVTVDTSDNAAATTATPSGLHRRQKVLSVVFLVVLPYL 362
Query: 122 KSKLHSIYNK 131
KSKLH+IYN+
Sbjct: 363 KSKLHTIYNR 392
>BP065734
Length = 406
Score = 56.2 bits (134), Expect = 8e-09
Identities = 26/31 (83%), Positives = 28/31 (89%)
Frame = -2
Query: 242 IKTLQGALLSCTYTVLDYAQTGLIAAVFFFK 272
++ LQGALLSCTYTVLDYAQTGLIA FFFK
Sbjct: 303 LQPLQGALLSCTYTVLDYAQTGLIATYFFFK 211
>AV772031
Length = 599
Score = 48.1 bits (113), Expect = 2e-06
Identities = 23/31 (74%), Positives = 26/31 (83%)
Frame = -2
Query: 242 IKTLQGALLSCTYTVLDYAQTGLIAAVFFFK 272
++ LQGALLS TYTVLD AQTGLIA FF+K
Sbjct: 499 LQALQGALLSSTYTVLDNAQTGLIATYFFYK 407
>TC14940
Length = 781
Score = 42.0 bits (97), Expect = 2e-04
Identities = 24/79 (30%), Positives = 35/79 (43%)
Frame = +2
Query: 289 TVYPPPPPPPPPKVAKEGVQLPSDRTICPLCLQKRVNPSVMTVSGFVFCYACIFKFLTQY 348
TV PP PP P + CP+C+ V + T G +FC ACI ++
Sbjct: 104 TVKKPPEPPKEPVFS------------CPICMGPLVE-EMSTRCGHIFCKACIKAAISAQ 244
Query: 349 KRCPATMVPATVDQIRRLF 367
+CP TV ++ R+F
Sbjct: 245 NKCPTCRKKVTVKELIRVF 301
>BI418821
Length = 614
Score = 30.0 bits (66), Expect(2) = 5e-04
Identities = 12/24 (50%), Positives = 13/24 (54%)
Frame = -3
Query: 277 WYQSAEERMSAPTVYPPPPPPPPP 300
W A +S PPPPPPPPP
Sbjct: 435 WQSLARCPVSPQL*QPPPPPPPPP 364
Score = 29.3 bits (64), Expect(2) = 5e-04
Identities = 11/22 (50%), Positives = 13/22 (59%)
Frame = -3
Query: 292 PPPPPPPPPKVAKEGVQLPSDR 313
PPPPPPPPP + P+ R
Sbjct: 387 PPPPPPPPPLRLSPPLHPPNPR 322
>BP052725
Length = 334
Score = 31.2 bits (69), Expect(2) = 7e-04
Identities = 18/64 (28%), Positives = 24/64 (37%), Gaps = 4/64 (6%)
Frame = -3
Query: 241 WIKTLQGALLSCTYTVLDYAQTGLIAAVFFFKMMEWWYQSAEERMSAPTVY----PPPPP 296
W+ +Q +SC Y F F + + S + T Y P PPP
Sbjct: 203 WLCEVQNYYISCPY--------------FMFVLFSHFLSSGKMNFQFETTYMIIFPRPPP 66
Query: 297 PPPP 300
PPPP
Sbjct: 65 PPPP 54
Score = 27.7 bits (60), Expect(2) = 7e-04
Identities = 10/18 (55%), Positives = 12/18 (66%)
Frame = -2
Query: 293 PPPPPPPPKVAKEGVQLP 310
PPPPPPPP + + V P
Sbjct: 78 PPPPPPPPSLLEVCVYKP 25
>TC8345
Length = 607
Score = 39.7 bits (91), Expect = 8e-04
Identities = 17/24 (70%), Positives = 19/24 (78%)
Frame = +1
Query: 272 KMMEWWYQSAEERMSAPTVYPPPP 295
+MME WYQS EER+SA TV PP P
Sbjct: 13 EMMECWYQSVEERISASTVCPPSP 84
>TC11625 weakly similar to UP|O81015 (O81015) At2g26920 protein, partial
(8%)
Length = 410
Score = 36.2 bits (82), Expect = 0.009
Identities = 18/39 (46%), Positives = 18/39 (46%)
Frame = -3
Query: 280 SAEERMSAPTVYPPPPPPPPPKVAKEGVQLPSDRTICPL 318
S R S TV PPPPPPPPP L T PL
Sbjct: 333 SLHRRFSHSTVIPPPPPPPPPPPPPSTTLLSLSETPGPL 217
>TC7926 homologue to UP|PGKH_TOBAC (Q42961) Phosphoglycerate kinase,
chloroplast precursor , partial (86%)
Length = 1857
Score = 35.8 bits (81), Expect = 0.012
Identities = 13/32 (40%), Positives = 18/32 (55%)
Frame = +2
Query: 269 FFFKMMEWWYQSAEERMSAPTVYPPPPPPPPP 300
F +EW + + +S + PPPPPPPPP
Sbjct: 101 FLSLSLEWHQRQQHQPLSLSSAPPPPPPPPPP 196
Score = 30.4 bits (67), Expect = 0.49
Identities = 14/35 (40%), Positives = 18/35 (51%), Gaps = 4/35 (11%)
Frame = +2
Query: 270 FFKMMEWWYQSAEER----MSAPTVYPPPPPPPPP 300
F + W+Q + + SAP PPPPPP PP
Sbjct: 101 FLSLSLEWHQRQQHQPLSLSSAPPPPPPPPPPHPP 205
>AV425903
Length = 375
Score = 34.7 bits (78), Expect = 0.026
Identities = 17/44 (38%), Positives = 24/44 (53%), Gaps = 3/44 (6%)
Frame = +1
Query: 268 VFFFKMMEWWYQSAEER---MSAPTVYPPPPPPPPPKVAKEGVQ 308
+ + ++ W Y R ++APT PPPPP PPP A GV+
Sbjct: 40 IISYHIISWAYVFPPPRSAALTAPTTTPPPPPMPPP-TANNGVR 168
>TC8581 similar to UP|Q94II4 (Q94II4) NAM-like protein, partial (9%)
Length = 1158
Score = 34.3 bits (77), Expect = 0.034
Identities = 11/13 (84%), Positives = 13/13 (99%)
Frame = -3
Query: 289 TVYPPPPPPPPPK 301
++YPPPPPPPPPK
Sbjct: 379 SLYPPPPPPPPPK 341
Score = 27.3 bits (59), Expect = 4.2
Identities = 10/23 (43%), Positives = 12/23 (51%)
Frame = -3
Query: 277 WYQSAEERMSAPTVYPPPPPPPP 299
W++ E PPPPPPPP
Sbjct: 412 WHRFGSEGT*RSLYPPPPPPPPP 344
>TC19962
Length = 374
Score = 26.9 bits (58), Expect(2) = 0.042
Identities = 9/20 (45%), Positives = 13/20 (65%)
Frame = -2
Query: 280 SAEERMSAPTVYPPPPPPPP 299
S+ ++ P+ PPPPPPP
Sbjct: 190 SSHSHLTPPSPANPPPPPPP 131
Score = 25.8 bits (55), Expect(2) = 0.042
Identities = 13/33 (39%), Positives = 16/33 (48%)
Frame = -2
Query: 294 PPPPPPPKVAKEGVQLPSDRTICPLCLQKRVNP 326
PPPPPPP + L + PLC+ V P
Sbjct: 151 PPPPPPPNITT--APL*RSKPTPPLCVSL*VWP 59
>TC11036 similar to UP|COQ4_ARATH (Q9ZPR0) Ubiquinone biosynthesis protein
COQ4 homolog, partial (93%)
Length = 992
Score = 33.9 bits (76), Expect = 0.044
Identities = 15/29 (51%), Positives = 18/29 (61%), Gaps = 5/29 (17%)
Frame = -3
Query: 280 SAEERMS-----APTVYPPPPPPPPPKVA 303
S +ER+S PT +PPP PPPPP A
Sbjct: 270 SHQERLSNPLSEGPTTHPPPSPPPPPPAA 184
>TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor kinase 1;1,
partial (16%)
Length = 486
Score = 29.3 bits (64), Expect(2) = 0.068
Identities = 9/22 (40%), Positives = 14/22 (62%)
Frame = +2
Query: 277 WYQSAEERMSAPTVYPPPPPPP 298
+YQ++ + + PPPPPPP
Sbjct: 2 YYQTSPHHVGCSSTLPPPPPPP 67
Score = 22.7 bits (47), Expect(2) = 0.068
Identities = 10/23 (43%), Positives = 11/23 (47%)
Frame = +1
Query: 295 PPPPPPKVAKEGVQLPSDRTICP 317
PPP P KV + PS T P
Sbjct: 139 PPPSPTKVTENPPTAPSTSTKSP 207
>AV411356
Length = 358
Score = 33.1 bits (74), Expect = 0.076
Identities = 15/55 (27%), Positives = 28/55 (50%), Gaps = 3/55 (5%)
Frame = +2
Query: 316 CPLCLQKRVNPSVMTVSGFVFCYACIFKFL---TQYKRCPATMVPATVDQIRRLF 367
C +CL P V+T G +FC+ C++++L + K CP T+ + ++
Sbjct: 44 CNICLDLAREP-VVTCCGHLFCWTCVYRWLHLHSDAKECPVCKGEVTLKSVTPIY 205
>TC14669 similar to PIR|E86326|E86326 protein F18O14.3 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(77%)
Length = 1120
Score = 32.7 bits (73), Expect = 0.099
Identities = 17/59 (28%), Positives = 30/59 (50%), Gaps = 3/59 (5%)
Frame = +2
Query: 297 PPPPKVAKEGVQLPSDRTICPLCLQKRVNPSVMTVSGFVFCYACIFKFL---TQYKRCP 352
PP P A + C +C +P ++T+ G +FC+ C++K+L +Q + CP
Sbjct: 74 PPSPSYAGNN-NSDAGNFECNICFDLAQDP-IITLCGHLFCWPCLYKWLHFHSQSRECP 244
>TC10567 similar to UP|AAR99376 (AAR99376) Ring domain containing protein,
partial (27%)
Length = 423
Score = 32.7 bits (73), Expect = 0.099
Identities = 13/30 (43%), Positives = 21/30 (69%)
Frame = +2
Query: 316 CPLCLQKRVNPSVMTVSGFVFCYACIFKFL 345
C +CL RV V+T+ G ++C+ CI+K+L
Sbjct: 86 CSICLD-RVQDPVVTLCGHLYCWPCIYKWL 172
>TC14097 homologue to UP|GCSP_PEA (P26969) Glycine dehydrogenase
[decarboxylating], mitochondrial precursor (Glycine
decarboxylase) (Glycine cleavage system P-protein) ,
partial (97%)
Length = 3660
Score = 27.3 bits (59), Expect(2) = 0.12
Identities = 10/24 (41%), Positives = 11/24 (45%)
Frame = +3
Query: 277 WYQSAEERMSAPTVYPPPPPPPPP 300
W+ S PPPPPPPP
Sbjct: 165 WFPKRSTTESMNRCGTPPPPPPPP 236
Score = 23.5 bits (49), Expect(2) = 0.12
Identities = 12/26 (46%), Positives = 13/26 (49%)
Frame = +3
Query: 295 PPPPPPKVAKEGVQLPSDRTICPLCL 320
PPPPPP + LP CPL L
Sbjct: 213 PPPPPPPPCRFTPLLPPG--TCPLFL 284
>TC15380 similar to UP|Q8H5P5 (Q8H5P5) OJ1003_C06.27 protein, partial (28%)
Length = 1306
Score = 32.3 bits (72), Expect = 0.13
Identities = 16/43 (37%), Positives = 21/43 (48%)
Frame = +2
Query: 293 PPPPPPPPKVAKEGVQLPSDRTICPLCLQKRVNPSVMTVSGFV 335
PPPPPPPP+ S + P CL++R N + S V
Sbjct: 299 PPPPPPPPRPFPS--LCSSTAEVSPSCLRRRSNSTPSAASSAV 421
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.324 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,218,045
Number of Sequences: 28460
Number of extensions: 136776
Number of successful extensions: 8002
Number of sequences better than 10.0: 392
Number of HSP's better than 10.0 without gapping: 2775
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5155
length of query: 370
length of database: 4,897,600
effective HSP length: 92
effective length of query: 278
effective length of database: 2,279,280
effective search space: 633639840
effective search space used: 633639840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC137603.5


