

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
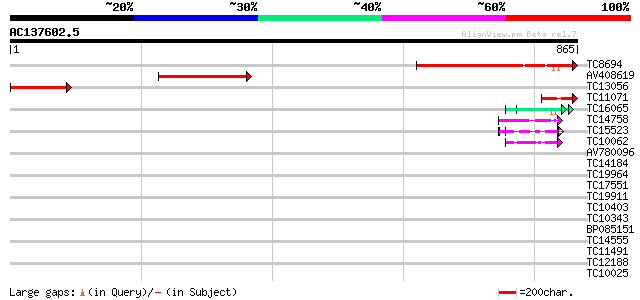
Query= AC137602.5 - phase: 0
(865 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8694 similar to UP|IF38_MEDTR (Q9XHM1) Eukaryotic translation ... 382 e-106
AV408619 258 2e-69
TC13056 similar to UP|IF38_MEDTR (Q9XHM1) Eukaryotic translation... 154 4e-38
TC11071 similar to UP|IF38_MEDTR (Q9XHM1) Eukaryotic translation... 50 2e-06
TC16065 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich... 48 6e-06
TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding p... 47 1e-05
TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-bindi... 46 2e-05
TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein... 45 6e-05
AV780096 41 0.001
TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-l... 39 0.003
TC19964 similar to UP|Q40363 (Q40363) NuM1 protein, partial (11%) 39 0.004
TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, part... 38 0.008
TC19911 weakly similar to PIR|A44805|A44805 eggshell protein pre... 38 0.008
TC10403 similar to UP|O80815 (O80815) T8F5.22 protein, partial (... 37 0.013
TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (... 36 0.030
BP085151 35 0.039
TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precu... 35 0.066
TC11491 weakly similar to UP|EBN1_EBV (P03211) EBNA-1 nuclear pr... 35 0.066
TC12188 similar to GB|AAF48767.2|22832753|AE003506 CG6474-PA {Dr... 34 0.087
TC10025 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein... 34 0.11
>TC8694 similar to UP|IF38_MEDTR (Q9XHM1) Eukaryotic translation initiation
factor 3 subunit 8 (eIF3 p110) (eIF3c), partial (23%)
Length = 986
Score = 382 bits (982), Expect = e-106
Identities = 203/257 (78%), Positives = 221/257 (85%), Gaps = 12/257 (4%)
Frame = +2
Query: 621 AATRVLINGDFQKAFDIIASLEVWKFVKNRDTVLEMLKDKIKEEALRTYLFTFSSSYDSL 680
AATRVL GD+ KAF+II SL+VWKFVKNRDTVLEMLKDKIKEEALRTYLFTFSSSYDSL
Sbjct: 14 AATRVLSKGDYNKAFEIIVSLDVWKFVKNRDTVLEMLKDKIKEEALRTYLFTFSSSYDSL 193
Query: 681 SVDQLTNFFDLSLPRAHSIVSRMMINEELHASWDQPTGCIIFRNVELSRVQALAFQLTEK 740
SVDQLT FDLS+PR HSIVS+MM+NEELHASWDQP+GCI+F+NVELSR+QAL FQLTEK
Sbjct: 194 SVDQLTKIFDLSVPRIHSIVSKMMVNEELHASWDQPSGCIVFQNVELSRLQALTFQLTEK 373
Query: 741 LSILAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQ 800
LSI AESNERA+EAR+GGGGLDLPPRRRDGQDYAAAAAGGGGG +GGRWQDLS SQ RQ
Sbjct: 374 LSIFAESNERAAEARIGGGGLDLPPRRRDGQDYAAAAAGGGGG--AGGRWQDLSLSQPRQ 547
Query: 801 GSGRAGY---GGRALSFNQAGGSGGYSRGR----GMGGGGYQN----SSRT-QGGSALRG 848
GSGRAGY GGR +FNQA +GGYSR R G GGGGYQ+ S R QGGSALRG
Sbjct: 548 GSGRAGYGSGGGRPFAFNQA--AGGYSRDRTGRGGGGGGGYQSQYGGSGRAHQGGSALRG 721
Query: 849 PHGDVSTRMVSLRGVRA 865
HGD S+RMVSL+G RA
Sbjct: 722 HHGDGSSRMVSLKGARA 772
>AV408619
Length = 427
Score = 258 bits (660), Expect = 2e-69
Identities = 123/142 (86%), Positives = 135/142 (94%)
Frame = +1
Query: 227 QILFSVVSAQFDVNPGLSGHMPINVWKKCVQNMLVILDILVQHPNIKVDDSVELDENESK 286
+ILFSVVSAQFDVNPGL+GHMPINVWKKCV NML+ILD+LVQ+PNI VDDSVELDE E++
Sbjct: 1 EILFSVVSAQFDVNPGLNGHMPINVWKKCVHNMLIILDVLVQYPNIMVDDSVELDEAETQ 180
Query: 287 KGDDYDGPIHVWGNLVAFLEKIDAEFFKSLQCIDPHTREYVERLRDEPLFVVLAQNVQEY 346
KG DYDGPI VWGNLVAFLE+IDAEFFKSLQCIDPHTREYVERLRDEPLF+VLAQNVQEY
Sbjct: 181 KGSDYDGPIRVWGNLVAFLERIDAEFFKSLQCIDPHTREYVERLRDEPLFLVLAQNVQEY 360
Query: 347 LESIGDFKASSKVALKRVELIY 368
LE GDFKA+SKVAL+RVEL+Y
Sbjct: 361 LERTGDFKAASKVALRRVELVY 426
>TC13056 similar to UP|IF38_MEDTR (Q9XHM1) Eukaryotic translation initiation
factor 3 subunit 8 (eIF3 p110) (eIF3c), partial (12%)
Length = 607
Score = 154 bits (390), Expect = 4e-38
Identities = 76/94 (80%), Positives = 86/94 (90%)
Frame = +2
Query: 1 MASTVDQIKNAIKINDWVSLQETFDKINKQLDKVMRVIESQKIPNLYIKALVMLEDFLAQ 60
MA+TVDQ+KNA+KINDWVSLQE+FDKINKQL+KVMRV S K P +YIKALVMLEDFLA+
Sbjct: 326 MAATVDQMKNAVKINDWVSLQESFDKINKQLEKVMRVTGSDKAPRIYIKALVMLEDFLAE 505
Query: 61 ASSNKDAKKKMSPTNAKAFNSMKQKLKKNNKQYE 94
A +NKDAKKKMS +NAKA N+MKQKLKKNNKQYE
Sbjct: 506 ALANKDAKKKMSSSNAKALNAMKQKLKKNNKQYE 607
>TC11071 similar to UP|IF38_MEDTR (Q9XHM1) Eukaryotic translation initiation
factor 3 subunit 8 (eIF3 p110) (eIF3c), partial (3%)
Length = 552
Score = 49.7 bits (117), Expect = 2e-06
Identities = 28/54 (51%), Positives = 33/54 (60%)
Frame = +1
Query: 812 LSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGDVSTRMVSLRGVRA 865
L + GG+GG + GG G + GGS LRG GDVSTRMVSL+GVRA
Sbjct: 106 LGIERDGGTGGGYQSAYSGGSGRAH----HGGSTLRGHQGDVSTRMVSLKGVRA 255
Score = 40.8 bits (94), Expect = 0.001
Identities = 23/39 (58%), Positives = 26/39 (65%), Gaps = 3/39 (7%)
Frame = +3
Query: 792 DLSYSQTRQGSGRAGY---GGRALSFNQAGGSGGYSRGR 827
DLS SQ QG GR GY GG ++F+QA GS GYSR R
Sbjct: 3 DLSLSQPXQGGGRTGYGSGGGXPMAFSQAAGS-GYSRDR 116
>TC16065 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich
glycoprotein DZ-HRGP precursor, partial (20%)
Length = 608
Score = 48.1 bits (113), Expect = 6e-06
Identities = 34/99 (34%), Positives = 39/99 (39%), Gaps = 6/99 (6%)
Frame = -1
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRW---QDLSYSQTRQGSGRAGYGGRALS 813
GG G PP+ G Y G GG GG Q QG GR G +
Sbjct: 347 GGRGYGGPPQGGRGGGYGGGGGPGYGGGGRGGHGVPQQQYGGPPEYQGRGRGGPSQQGGR 168
Query: 814 FNQAGGSGGYSRGRGMGGGGY---QNSSRTQGGSALRGP 849
+GG GGY G G GGGG + S + GG R P
Sbjct: 167 GGYSGGGGGYGGGGGRGGGGMGSGRGVSPSHGGPPSRPP 51
Score = 47.0 bits (110), Expect = 1e-05
Identities = 39/105 (37%), Positives = 41/105 (38%), Gaps = 19/105 (18%)
Frame = -1
Query: 774 AAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYS--------- 824
AAAA GGGG GGR Y QG GYGG GG GG+
Sbjct: 386 AAAAPGGGGPGPQGGR----GYGGPPQGGRGGGYGGGGGPGYGGGGRGGHGVPQQQYGGP 219
Query: 825 ---RGRGMGG-------GGYQNSSRTQGGSALRGPHGDVSTRMVS 859
+GRG GG GGY GG RG G S R VS
Sbjct: 218 PEYQGRGRGGPSQQGGRGGYSGGGGGYGGGGGRGGGGMGSGRGVS 84
>TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding protein 2,
partial (93%)
Length = 725
Score = 47.0 bits (110), Expect = 1e-05
Identities = 37/99 (37%), Positives = 41/99 (41%), Gaps = 2/99 (2%)
Frame = +1
Query: 747 SNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGR-- 804
S E A G G +L R + A GGGGG GG + G GR
Sbjct: 232 SEEAMRSAIEGMNGNELDGRNITVNEAQARGGGGGGGGRGGGGYGG--------GGGRRE 387
Query: 805 AGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
GYGG S GG GGY G GGGGY R +GG
Sbjct: 388 GGYGGGGGSRGYGGGGGGYGGG---GGGGY--GGRREGG 489
Score = 38.9 bits (89), Expect = 0.004
Identities = 32/78 (41%), Positives = 33/78 (42%), Gaps = 1/78 (1%)
Frame = +1
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG RR+G GGGG GG G G GYGGR
Sbjct: 349 GGGGYGGGGGRREG-----GYGGGGGSRGYGGGGGGYG------GGGGGGYGGR--REGG 489
Query: 817 AGGSGGYSR-GRGMGGGG 833
GG GG SR G G GGGG
Sbjct: 490 YGGDGGGSRYGSGGGGGG 543
>TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-binding,
abscisic acid-inducible protein, complete
Length = 972
Score = 46.2 bits (108), Expect = 2e-05
Identities = 34/97 (35%), Positives = 36/97 (37%)
Frame = +2
Query: 748 NERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGY 807
NE + R GGGG G Y GG G G R+ G G GY
Sbjct: 578 NEAQARGRGGGGG---------GGGYGGGRREGGYGGGGGSRYS---------GGGGGGY 703
Query: 808 GGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGS 844
GG G GGY R GGGGY S GGS
Sbjct: 704 GG--------SGGGGYGGRREGGGGGYGGSREGGGGS 790
Score = 44.3 bits (103), Expect = 8e-05
Identities = 37/88 (42%), Positives = 40/88 (45%)
Frame = +2
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG RR+G GGGG SGG GSG GYGGR +
Sbjct: 608 GGGGGGYGGGRREG-----GYGGGGGSRYSGGGGGGYG------GSGGGGYGGR-----R 739
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGS 844
GG GGY G GGGG + SS GGS
Sbjct: 740 EGGGGGYG-GSREGGGGSRYSS---GGS 811
Score = 43.9 bits (102), Expect = 1e-04
Identities = 36/99 (36%), Positives = 42/99 (42%), Gaps = 2/99 (2%)
Frame = +2
Query: 747 SNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGR-- 804
S + +A G G D+ R + A GGGGG GG + G GR
Sbjct: 506 SEQSMKDAIEGMNGQDIDGRNVTVNEAQARGRGGGGG---GGGY----------GGGRRE 646
Query: 805 AGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
GYGG S GG GGY G GGGGY R +GG
Sbjct: 647 GGYGGGGGSRYSGGGGGGYG---GSGGGGY--GGRREGG 748
Score = 30.8 bits (68), Expect = 0.96
Identities = 18/40 (45%), Positives = 21/40 (52%), Gaps = 1/40 (2%)
Frame = +2
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGT-SSGGRWQDLSY 795
GGGG R G Y + GGGG SSGG W+D S+
Sbjct: 713 GGGGYG-GRREGGGGGYGGSREGGGGSRYSSGGSWRD*SF 829
Score = 28.1 bits (61), Expect = 6.2
Identities = 19/54 (35%), Positives = 23/54 (42%), Gaps = 2/54 (3%)
Frame = +2
Query: 800 QGSGRAGYGGRALSFN--QAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
+G GR ++ N QA G GG G G GGG + GGS G G
Sbjct: 533 EGMNGQDIDGRNVTVNEAQARGRGGGGGGGGYGGGRREGGYGGGGGSRYSGGGG 694
>TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein 2, partial
(66%)
Length = 481
Score = 44.7 bits (104), Expect = 6e-05
Identities = 34/87 (39%), Positives = 38/87 (43%)
Frame = +3
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
G G + RR G A G GGG GG + Y +G G GYGG
Sbjct: 243 GPNGQAVQGTRRGG---GAGGGGRGGGGYGGGGYGGGGYGGGGRGGG--GYGGGG----G 395
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGG 843
GGSGGY G G GGGG + R GG
Sbjct: 396 YGGSGGYGGGGGYGGGG-RGGGRGGGG 473
Score = 40.8 bits (94), Expect = 0.001
Identities = 35/107 (32%), Positives = 40/107 (36%), Gaps = 4/107 (3%)
Frame = +3
Query: 731 QALAFQLTEKLSILAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGG----GTSS 786
+A A ++T + R A GGGG R G Y GGGG G
Sbjct: 219 RAKAIEVTGPNGQAVQGTRRGGGA--GGGG-------RGGGGYGGGGYGGGGYGGGGRGG 371
Query: 787 GGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGG 833
GG G GYGG + GG GG RG G GGGG
Sbjct: 372 GGY------------GGGGGYGGSG-GYGGGGGYGGGGRGGGRGGGG 473
Score = 38.5 bits (88), Expect = 0.005
Identities = 26/71 (36%), Positives = 26/71 (36%)
Frame = +3
Query: 781 GGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRT 840
GGG GGR G G GYGG GG GGY G G GG G
Sbjct: 279 GGGAGGGGRGGG--------GYGGGGYGGGGYGGGGRGG-GGYGGGGGYGGSGGYGGGGG 431
Query: 841 QGGSALRGPHG 851
GG G G
Sbjct: 432 YGGGGRGGGRG 464
Score = 37.7 bits (86), Expect = 0.008
Identities = 31/83 (37%), Positives = 34/83 (40%)
Frame = +3
Query: 769 DGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRG 828
DG+ A G G G TR+G G AG GGR G GGY G G
Sbjct: 213 DGRAKAIEVTGPNGQAVQG----------TRRGGG-AGGGGR--------GGGGYG-GGG 332
Query: 829 MGGGGYQNSSRTQGGSALRGPHG 851
GGGGY R GG G +G
Sbjct: 333 YGGGGYGGGGRGGGGYGGGGGYG 401
Score = 32.3 bits (72), Expect = 0.33
Identities = 30/77 (38%), Positives = 30/77 (38%)
Frame = +3
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG R G Y GGGG SGG G GYGG
Sbjct: 351 GGGG-------RGGGGYG----GGGGYGGSGGY------------GGGGGYGG------- 440
Query: 817 AGGSGGYSRGRGMGGGG 833
GG GG GRG GGGG
Sbjct: 441 -GGRGG---GRGGGGGG 479
>AV780096
Length = 564
Score = 40.8 bits (94), Expect = 0.001
Identities = 40/158 (25%), Positives = 64/158 (40%), Gaps = 20/158 (12%)
Frame = +1
Query: 88 KNNKQYEDLIIKFRETPQSEEEKFEDDEDSDQYGSDDDEIIEPDQLKKPEPVSDSETSEL 147
+ N D+ + + EEE+ +++E DQ DD+E+ + + + D
Sbjct: 4 ERNLDLRDMARRNGSDSEEEEEEQQEEEVHDQIEEDDEEL---EAVARSASSDDEPDESG 174
Query: 148 GNDRPGDDDDAPWDQ--------KLRKKDRLLEKMFMKKPSEITWDTVNNKFKEILEAR- 198
G D DDD D+ R+K RL E +KK K +EIL+A+
Sbjct: 175 GEDAVAADDDNQDDEGNAADPEISKREKARLKELQQLKK----------QKIQEILDAQN 324
Query: 199 -------GRKGTGR----FEQVEQFTFLTKVAKTPAQK 225
+G GR +Q E F L K ++ +QK
Sbjct: 325 AAIDADMNNRGKGRLNYLLQQTELFAHLAKADQSSSQK 438
>TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-like
protein, partial (57%)
Length = 1141
Score = 39.3 bits (90), Expect = 0.003
Identities = 32/90 (35%), Positives = 38/90 (41%)
Frame = +3
Query: 747 SNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAG 806
S+ E GGGG D + + G G G + GG YS T G G G
Sbjct: 276 SSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGG------YS-TGGGYGTGG 434
Query: 807 YGGRALSFNQAGGSGGYSRGRGMGGGGYQN 836
GG ++ GG GGYS G GGGY N
Sbjct: 435 GGGG--EYSTGGGGGGYSTG----GGGYGN 506
Score = 37.0 bits (84), Expect = 0.013
Identities = 27/102 (26%), Positives = 36/102 (34%), Gaps = 5/102 (4%)
Frame = +3
Query: 746 ESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRA 805
+S + S GGG + + +GGGG S + +YS G
Sbjct: 207 DSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSG---YNKTAYSTDEPSGGGY 377
Query: 806 GYGGRALSFNQ-----AGGSGGYSRGRGMGGGGYQNSSRTQG 842
G GG ++ GG GG G GGGGY G
Sbjct: 378 GTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYG 503
>TC19964 similar to UP|Q40363 (Q40363) NuM1 protein, partial (11%)
Length = 562
Score = 38.9 bits (89), Expect = 0.004
Identities = 28/75 (37%), Positives = 34/75 (45%), Gaps = 1/75 (1%)
Frame = +2
Query: 753 EARLGGGGLDLPPRR-RDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRA 811
++ LGG L + + RD Q + GG G GGR+ S G GR G GGR
Sbjct: 77 DSDLGGFTLAVDEAKPRDNQGSGGRSGGGRSGGGRGGRFD----SGRGGGRGRFGSGGRG 244
Query: 812 LSFNQAGGSGGYSRG 826
GGSGG RG
Sbjct: 245 GDRGGRGGSGGGGRG 289
Score = 28.1 bits (61), Expect = 6.2
Identities = 23/61 (37%), Positives = 29/61 (46%), Gaps = 1/61 (1%)
Frame = +2
Query: 792 DLSYSQTRQGSGRAGYGGRALSFNQAGGSGG-YSRGRGMGGGGYQNSSRTQGGSALRGPH 850
D + + QGSG GGR+ GG GG + GRG G G + + R GG RG
Sbjct: 110 DEAKPRDNQGSGGRSGGGRS-----GGGRGGRFDSGRGGGRGRFGSGGR--GGD--RGGR 262
Query: 851 G 851
G
Sbjct: 263 G 265
>TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, partial (22%)
Length = 505
Score = 37.7 bits (86), Expect = 0.008
Identities = 30/95 (31%), Positives = 38/95 (39%)
Frame = -3
Query: 749 ERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYG 808
+R + R G G R G+ + G G G W+ S+ R G GR+G G
Sbjct: 431 KRGARGRGGSG-------RSRGRRRCSRGGRGRGSIWRRGGWRRRGRSRWR*GRGRSGGG 273
Query: 809 GRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
G GG G SRG G GG R +GG
Sbjct: 272 GGG------GGGGRRSRGSGGGGTNGGRRGRGKGG 186
>TC19911 weakly similar to PIR|A44805|A44805 eggshell protein precursor -
fluke (Schistosoma haematobium) {Schistosoma
haematobium;} , partial (24%)
Length = 350
Score = 37.7 bits (86), Expect = 0.008
Identities = 33/105 (31%), Positives = 43/105 (40%)
Frame = +1
Query: 748 NERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGY 807
N + GGG R D + + G G S GG + D + S R G G G
Sbjct: 16 NSAGNTGHSGGGSHGNDHRHSDNEHGGRRSETTGHGLS-GGGYDDDNRSGGRTGGG-YGD 189
Query: 808 GGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGD 852
GG + AGG G + G GGGGY + + G G +GD
Sbjct: 190 GG-----HSAGGYGDHGGRSGGGGGGYGDGGLSSGS----GGYGD 297
>TC10403 similar to UP|O80815 (O80815) T8F5.22 protein, partial (15%)
Length = 1392
Score = 37.0 bits (84), Expect = 0.013
Identities = 23/73 (31%), Positives = 30/73 (40%), Gaps = 5/73 (6%)
Frame = +3
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQG-----SGRAGYGGRALSFNQAGGSGGYS 824
G A+ G GG++S G W+ SY + R +GRA Y G
Sbjct: 645 GGSSGASVGSGWGGSNSEGGWKGYSYDRDRSSTPGSRTGRADYRNNGNRDEHPSGVPRPY 824
Query: 825 RGRGMGGGGYQNS 837
GRG G G Y +S
Sbjct: 825 GGRGRGRGSYSSS 863
>TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (Fibrillarin
2) (AtFib2), partial (85%)
Length = 1116
Score = 35.8 bits (81), Expect = 0.030
Identities = 27/77 (35%), Positives = 30/77 (38%), Gaps = 1/77 (1%)
Frame = +1
Query: 764 PPRRRDGQD-YAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGG 822
PPR R G + G G GGR +G GR G GGR GG
Sbjct: 40 PPRGRGGGGGFRGRGDRGRGRGDGGGRGGGRGTPFKARGGGRGGGGGR----------GG 189
Query: 823 YSRGRGMGGGGYQNSSR 839
GRG GGG + SR
Sbjct: 190 RGGGRGGRGGGMKGGSR 240
>BP085151
Length = 360
Score = 35.4 bits (80), Expect = 0.039
Identities = 31/83 (37%), Positives = 35/83 (41%), Gaps = 7/83 (8%)
Frame = -1
Query: 776 AAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGY--GGRALSFNQAGGSGGYSRGRGMGG-- 831
A A G GG +GGR GRAG GGRA G +GG GR GG
Sbjct: 309 AYAAGRGGLGAGGR-----------AGGRAGVRAGGRA-----GGRAGGREGGRAAGGRE 178
Query: 832 ---GGYQNSSRTQGGSALRGPHG 851
G + R +GG RGP G
Sbjct: 177 GGRAGARGVRRGRGGRRGRGP*G 109
>TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (43%)
Length = 1038
Score = 34.7 bits (78), Expect = 0.066
Identities = 32/107 (29%), Positives = 40/107 (36%)
Frame = -1
Query: 747 SNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAG 806
S+E A E R G L P DG+ A G G + DL ++ G+G
Sbjct: 663 SSEAAGE-RAGEREERLGPGASDGESETTGANAGAGAGAG-----DLLFTLAGAGAGEVA 502
Query: 807 YGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGDV 853
G + A G GG + G GGG GG L G G V
Sbjct: 501 GTGVSAGGGVAAGGGGVAGGGEKGGG-------EAGGGVLAGAGGGV 382
>TC11491 weakly similar to UP|EBN1_EBV (P03211) EBNA-1 nuclear protein,
partial (4%)
Length = 473
Score = 34.7 bits (78), Expect = 0.066
Identities = 22/53 (41%), Positives = 27/53 (50%), Gaps = 4/53 (7%)
Frame = +2
Query: 800 QGSGRAGYGGRAL----SFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRG 848
+G G G+GGR+ N+ G GG RG G G GYQ R +GGS G
Sbjct: 20 RGRGNYGFGGRSFVRVDYGNRGGDFGGRGRGPGGHGEGYQ-QGRGRGGSGRFG 175
Score = 32.0 bits (71), Expect = 0.43
Identities = 22/61 (36%), Positives = 29/61 (47%), Gaps = 1/61 (1%)
Frame = +2
Query: 780 GGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSG-GYSRGRGMGGGGYQNSS 838
G G GGR S+ + G+ +GGR GG G GY +GRG GG G ++
Sbjct: 23 GRGNYGFGGR----SFVRVDYGNRGGDFGGRGRG---PGGHGEGYQQGRGRGGSGRFGAT 181
Query: 839 R 839
R
Sbjct: 182 R 184
>TC12188 similar to GB|AAF48767.2|22832753|AE003506 CG6474-PA {Drosophila
melanogaster;} , partial (6%)
Length = 297
Score = 34.3 bits (77), Expect = 0.087
Identities = 28/71 (39%), Positives = 30/71 (41%), Gaps = 9/71 (12%)
Frame = +2
Query: 766 RRRDGQ--DYAAAAAGGGGGTSSGG---RWQDLSYSQTRQGSGRAGYGGRALSFN----Q 816
RRR G D AAGGGGG SGG W+ S GYG R + Q
Sbjct: 23 RRRVGARVDNRGLAAGGGGGGGSGGVRCHWRRARGSGDHHQHPEDGYGHRHHRADGHDEQ 202
Query: 817 AGGSGGYSRGR 827
G GG RGR
Sbjct: 203 REGGGGRGRGR 235
>TC10025 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein
(Fragment), partial (15%)
Length = 672
Score = 33.9 bits (76), Expect = 0.11
Identities = 18/74 (24%), Positives = 39/74 (52%)
Frame = +1
Query: 82 MKQKLKKNNKQYEDLIIKFRETPQSEEEKFEDDEDSDQYGSDDDEIIEPDQLKKPEPVSD 141
+K+ L K K+++ +K + ++ ++ E+ + G +DD+ + P + K+ V D
Sbjct: 193 VKEDLGKR-KRFQGKRLKKMSSDFESKKTGKEKEEDESAGHEDDKPVVPPKRKRSSKVKD 369
Query: 142 SETSELGNDRPGDD 155
E +L +PG+D
Sbjct: 370 EEEDDLFLRKPGND 411
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.134 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,175,867
Number of Sequences: 28460
Number of extensions: 151247
Number of successful extensions: 1860
Number of sequences better than 10.0: 133
Number of HSP's better than 10.0 without gapping: 1459
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1693
length of query: 865
length of database: 4,897,600
effective HSP length: 98
effective length of query: 767
effective length of database: 2,108,520
effective search space: 1617234840
effective search space used: 1617234840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC137602.5


