

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137546.2 - phase: 0
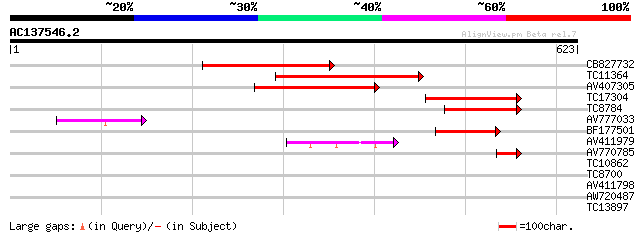
(623 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB827732 280 5e-76
TC11364 homologue to GB|CAA56354.1|510876|PVME1G NADP dependent ... 275 1e-74
AV407305 231 3e-61
TC17304 homologue to GB|CAA56354.1|510876|PVME1G NADP dependent ... 171 3e-43
TC8784 similar to UP|Q9M4Q9 (Q9M4Q9) NADP-dependent malic protei... 130 8e-31
AV777033 87 8e-18
BF177501 75 4e-14
AV411979 72 3e-13
AV770785 45 3e-05
TC10862 UP|RK2_LOTJA (Q9B1H9) Chloroplast 50S ribosomal protein ... 28 3.4
TC8700 28 4.4
AV411798 28 5.7
AW720487 27 9.8
TC13897 27 9.8
>CB827732
Length = 438
Score = 280 bits (716), Expect = 5e-76
Identities = 140/145 (96%), Positives = 144/145 (98%)
Frame = +2
Query: 212 ALYSALGGVRPSACLPVTIDVGTNNEKLLNDEFYIGLRQKRATGKEYYDLLHEFMTAVKQ 271
ALYSALGGVRPSACLPVTIDVGTNNE+LLNDEFYIGLRQKRATGKEYYDLLHEFMTAVKQ
Sbjct: 2 ALYSALGGVRPSACLPVTIDVGTNNEQLLNDEFYIGLRQKRATGKEYYDLLHEFMTAVKQ 181
Query: 272 NYGEKVLVQFEDFANHNAFELLAKYGTTHLVFNDDIQGTAAVVLAGVVASLKLIGGTLPE 331
NYGEKVLVQFEDFANHNAFELLAKYGTTHLVFNDDIQGTA+VVLAGVVA+LKLIGGTL +
Sbjct: 182 NYGEKVLVQFEDFANHNAFELLAKYGTTHLVFNDDIQGTASVVLAGVVAALKLIGGTLGD 361
Query: 332 HTFLFLGAGEAGTGIAELIALEMSK 356
HTFLFLGAGEAGTGIAELIALEMSK
Sbjct: 362 HTFLFLGAGEAGTGIAELIALEMSK 436
>TC11364 homologue to GB|CAA56354.1|510876|PVME1G NADP dependent malic
enzyme {Phaseolus vulgaris;} , partial (27%)
Length = 490
Score = 275 bits (704), Expect = 1e-74
Identities = 136/162 (83%), Positives = 150/162 (91%)
Frame = +1
Query: 293 LAKYGTTHLVFNDDIQGTAAVVLAGVVASLKLIGGTLPEHTFLFLGAGEAGTGIAELIAL 352
L +Y ++HLVFNDDIQGTA+VVLAG++ASLKL+GGTL +HTFLFLGAGEAGTGIAELIAL
Sbjct: 4 LDRYSSSHLVFNDDIQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIAL 183
Query: 353 EMSKQTKAPIEESRKKIWLVDSKGLIVSSRANSLQHFKKPWAHEHEPVSTLLDAVKIIKP 412
E+SKQTKAP+EE+RKK+WLVDSKGLIV SR SLQHFKKPWAHEHEPV LLDAVK IKP
Sbjct: 184 EISKQTKAPVEETRKKVWLVDSKGLIVRSRLESLQHFKKPWAHEHEPVKALLDAVKAIKP 363
Query: 413 TVLIGSSGVGKTFTKEVVEAMTEINKIPLILALSNPTSQSEC 454
TVLIGSSGVGKTFTKEVVEAM +N+ PLILALSNPTSQSEC
Sbjct: 364 TVLIGSSGVGKTFTKEVVEAMASLNEKPLILALSNPTSQSEC 489
>AV407305
Length = 414
Score = 231 bits (589), Expect = 3e-61
Identities = 112/137 (81%), Positives = 123/137 (89%)
Frame = +1
Query: 270 KQNYGEKVLVQFEDFANHNAFELLAKYGTTHLVFNDDIQGTAAVVLAGVVASLKLIGGTL 329
KQ YGEKVL+QFEDFANHNAF+LL KY +THLVFNDDIQGTA+VVLAG+VA+LKL+GG L
Sbjct: 4 KQTYGEKVLIQFEDFANHNAFDLLEKYRSTHLVFNDDIQGTASVVLAGLVAALKLVGGNL 183
Query: 330 PEHTFLFLGAGEAGTGIAELIALEMSKQTKAPIEESRKKIWLVDSKGLIVSSRANSLQHF 389
+H FLFLGAGEAGTGIAELIALE SKQT +P+EE RK IWLVDSKGLIVSSR SLQHF
Sbjct: 184 ADHRFLFLGAGEAGTGIAELIALETSKQTNSPLEEVRKNIWLVDSKGLIVSSRKESLQHF 363
Query: 390 KKPWAHEHEPVSTLLDA 406
KKPWAHEHEPV L+DA
Sbjct: 364 KKPWAHEHEPVKNLVDA 414
>TC17304 homologue to GB|CAA56354.1|510876|PVME1G NADP dependent malic
enzyme {Phaseolus vulgaris;} , partial (23%)
Length = 606
Score = 171 bits (433), Expect = 3e-43
Identities = 82/105 (78%), Positives = 95/105 (90%)
Frame = +2
Query: 458 EAYTWSEGRAIFASGSPFDPVEYKGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLL 517
EAYTWS+G+AIFASGSPFDPV+Y GK Y GQ+NNAYIFPGFGLG+++SGAIRV D+MLL
Sbjct: 2 EAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGLGLIISGAIRVRDEMLL 181
Query: 518 AASEALAKQVTEENYKKGLTYPPFSDIRKISANIAANVAAKAYEL 562
AASEALA QV++ENY KGL YPPF +IRKISA+IAA+VAAK YEL
Sbjct: 182 AASEALAAQVSQENYDKGLIYPPFPNIRKISAHIAASVAAKVYEL 316
>TC8784 similar to UP|Q9M4Q9 (Q9M4Q9) NADP-dependent malic protein (Malic
enzyme) , partial (18%)
Length = 729
Score = 130 bits (326), Expect = 8e-31
Identities = 62/85 (72%), Positives = 75/85 (87%)
Frame = +2
Query: 478 VEYKGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEENYKKGLT 537
V Y K + GQ+NNAYIFPGFGLG++MSG IRVHDD+LLAASEALA+QVT+E++ KGL
Sbjct: 2 VPYDDKVFVPGQANNAYIFPGFGLGLIMSGTIRVHDDLLLAASEALAEQVTQEHFDKGLI 181
Query: 538 YPPFSDIRKISANIAANVAAKAYEL 562
+PPF++IRKISA+IAA VAAKAYEL
Sbjct: 182 FPPFTNIRKISAHIAAKVAAKAYEL 256
>AV777033
Length = 592
Score = 87.0 bits (214), Expect = 8e-18
Identities = 47/109 (43%), Positives = 62/109 (56%), Gaps = 10/109 (9%)
Frame = +2
Query: 52 GYSLLRDPQYNKGLAFTEKERDAHYLRGLLPPTVSSQQLQEKKLMHNIRQYE-------- 103
G +L DP +NK F ERD LRGLLPP V S + Q + M++ R E
Sbjct: 263 GADILHDPWFNKDTGFPLTERDRLGLRGLLPPRVISFEQQYDRFMNSYRSLEKNTQCQPE 442
Query: 104 --VPLQKYVAMMDLQERNERLFYKLLIDNVEELLPIVYTPVVGEACQKY 150
V L K+ + L +RNE L+Y++LIDN++E PI+YTP VG CQ Y
Sbjct: 443 KVVSLAKWRILNRLHDRNETLYYRVLIDNIKEFAPIIYTPTVGLVCQNY 589
>BF177501
Length = 247
Score = 74.7 bits (182), Expect = 4e-14
Identities = 37/73 (50%), Positives = 50/73 (67%), Gaps = 1/73 (1%)
Frame = +1
Query: 468 IFASGSPFDPVEY-KGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQ 526
+FASGSPF+ V+ GK + Q+NN Y+FPG GLG ++SGA + D ML AASE LA
Sbjct: 25 VFASGSPFENVDLGNGKVGHVNQANNMYLFPGIGLGTLLSGARLITDGMLQAASECLASY 204
Query: 527 VTEENYKKGLTYP 539
+ E++ KG+ YP
Sbjct: 205 MVEDDILKGILYP 243
>AV411979
Length = 408
Score = 72.0 bits (175), Expect = 3e-13
Identities = 44/137 (32%), Positives = 74/137 (53%), Gaps = 14/137 (10%)
Frame = +1
Query: 305 DDIQGTAAVVLAGVVASLKLIGGTL---PEHTFLFLGAGEAGTGIAELIALEMSKQ---T 358
DD+QGTA V +AG++ +++ G + P+ + GAG AG G+ M++
Sbjct: 1 DDVQGTAGVAIAGLLGAVRAQGRPMIDFPKQKIVVAGAGSAGIGVLNAARKTMARMLGNN 180
Query: 359 KAPIEESRKKIWLVDSKGLIVSSRANSLQHFKKPWAHEHEPV--------STLLDAVKII 410
+A E ++ + W+VD+KGLI R N + P+A + + ++L++ VK +
Sbjct: 181 EAAFESAKSQFWVVDAKGLITEGREN-IDPDALPFARNLKEMDRQGLREGASLVEVVKQV 357
Query: 411 KPTVLIGSSGVGKTFTK 427
KP VL+G S VG F+K
Sbjct: 358 KPDVLLGLSAVGGLFSK 408
>AV770785
Length = 442
Score = 45.4 bits (106), Expect = 3e-05
Identities = 21/28 (75%), Positives = 25/28 (89%)
Frame = -1
Query: 535 GLTYPPFSDIRKISANIAANVAAKAYEL 562
GL +PPF+ +RKISA+IAA VAAKAYEL
Sbjct: 442 GLIFPPFTTLRKISAHIAAQVAAKAYEL 359
>TC10862 UP|RK2_LOTJA (Q9B1H9) Chloroplast 50S ribosomal protein L2,
complete
Length = 837
Score = 28.5 bits (62), Expect = 3.4
Identities = 19/68 (27%), Positives = 32/68 (46%)
Frame = +1
Query: 516 LLAASEALAKQVTEENYKKGLTYPPFSDIRKISANIAANVAAKAYELVCTATIGEQGQFT 575
L A+ A+AK + +E L P ++R IS N C+AT+G+ G
Sbjct: 466 LARAAGAVAKLIAKEGKSATLKLPS-GEVRLISKN-------------CSATVGQVGNVG 603
Query: 576 IDSEAFGR 583
++ ++ GR
Sbjct: 604 VNQKSLGR 627
>TC8700
Length = 607
Score = 28.1 bits (61), Expect = 4.4
Identities = 17/54 (31%), Positives = 28/54 (51%), Gaps = 9/54 (16%)
Frame = +3
Query: 141 PVVGEACQKYGSIYK---------RPQGLYISLKEKGKILEVLKNWPERSIQVI 185
P +G A K G +K +P I L +KG+ILEVL++ ++++ I
Sbjct: 87 PWLGNALPKVGFNFKQLHTSFAGWKPHATAIKLSDKGEILEVLEDCEGKTLKFI 248
>AV411798
Length = 397
Score = 27.7 bits (60), Expect = 5.7
Identities = 20/75 (26%), Positives = 32/75 (42%), Gaps = 8/75 (10%)
Frame = +3
Query: 134 LLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPERSIQVI-------- 185
L PIV C G+ P G+ + L ++ I+ L P IQ++
Sbjct: 168 LSPIVIFATNRGICNVRGTDMASPHGIPVDLLDRLAIVRTLIYGPAEMIQILAIRAQVEE 347
Query: 186 VVTDGERILGLGDLG 200
+V D E + LG++G
Sbjct: 348 LVVDEESLAFLGEIG 392
>AW720487
Length = 575
Score = 26.9 bits (58), Expect = 9.8
Identities = 22/84 (26%), Positives = 35/84 (41%)
Frame = -1
Query: 461 TWSEGRAIFASGSPFDPVEYKGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLLAAS 520
TWS GR + G PF PV+ + S + N I PG LG V + +
Sbjct: 278 TWSFGRR*TSPGCPFHPVDRNSR--LSPITGNP-ILPGGRLGAVNGDLCL*KREKRESDI 108
Query: 521 EALAKQVTEENYKKGLTYPPFSDI 544
A +T ++ ++PP+ +
Sbjct: 107 TTKAWTITSTSHSCSSSFPPYHSL 36
>TC13897
Length = 428
Score = 26.9 bits (58), Expect = 9.8
Identities = 12/31 (38%), Positives = 14/31 (44%)
Frame = +1
Query: 77 LRGLLPPTVSSQQLQEKKLMHNIRQYEVPLQ 107
L LPPT S K HN+ + VP Q
Sbjct: 91 LNPFLPPTPKSHYCSLKNASHNLHSFNVPSQ 183
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,547,355
Number of Sequences: 28460
Number of extensions: 118946
Number of successful extensions: 580
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 576
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 577
length of query: 623
length of database: 4,897,600
effective HSP length: 96
effective length of query: 527
effective length of database: 2,165,440
effective search space: 1141186880
effective search space used: 1141186880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC137546.2


