

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137510.9 + phase: 0
(354 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
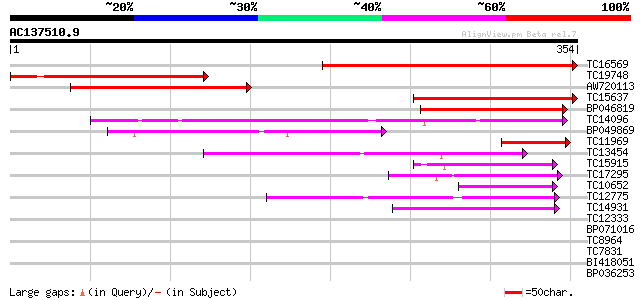
Score E
Sequences producing significant alignments: (bits) Value
TC16569 similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpyruv... 274 2e-74
TC19748 weakly similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoen... 169 6e-43
AW720113 135 1e-32
TC15637 similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpyruv... 135 1e-32
BP046819 107 2e-24
TC14096 homologue to UP|CPTR_PEA (P21727) Triose phosphate/phosp... 92 1e-19
BP049869 73 6e-14
TC11969 similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpyruv... 68 2e-12
TC13454 homologue to UP|O64910 (O64910) Glucose-6-phosphate/phos... 62 1e-10
TC15915 similar to UP|Q9SFE9 (Q9SFE9) T26F17.9 (GONST5 golgi nuc... 61 2e-10
TC17295 similar to UP|Q94JT2 (Q94JT2) AT5g17630/K10A8_110, parti... 55 2e-08
TC10652 similar to UP|Q9SFE9 (Q9SFE9) T26F17.9 (GONST5 golgi nuc... 54 3e-08
TC12775 similar to UP|Q9FG70 (Q9FG70) Gb|AAF04433.1, partial (45%) 46 8e-06
TC14931 similar to UP|Q9M9Z3 (Q9M9Z3) F4H5.5 protein, partial (55%) 46 1e-05
TC12333 similar to GB|AAP42755.1|30984584|BT008742 At2g30460 {Ar... 37 0.005
BP071016 30 0.47
TC8964 weakly similar to UP|Q94F54 (Q94F54) AT3g15070/K15M2_22, ... 28 1.8
TC7831 similar to UP|Q8H946 (Q8H946) Phosphoenolpyruvate carboxy... 27 4.0
BI418051 27 5.2
BP036253 27 6.8
>TC16569 similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpyruvate
translocator protein-like, partial (46%)
Length = 827
Score = 274 bits (700), Expect = 2e-74
Identities = 141/159 (88%), Positives = 151/159 (94%)
Frame = +3
Query: 196 FHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNLLLYMAPLAAMILLPVTLYIEG 255
FHLFGFLVCVGSTAGRALKSVVQGIILTSE+EKL+SMNLLLYMAP+AAMILLP TLYIEG
Sbjct: 3 FHLFGFLVCVGSTAGRALKSVVQGIILTSESEKLNSMNLLLYMAPMAAMILLPFTLYIEG 182
Query: 256 NVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVV 315
NV A+T+EK + D FIVFLL+GNATVAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVV
Sbjct: 183 NVAAVTLEKVKGDRFIVFLLVGNATVAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVV 362
Query: 316 SVLIFRNPVTVMGMTGFGITIMGVVLYSEAKKRSKGASH 354
SVLIFRNPVTVMG+TGF +TIMGV+LYSEAKKRSK SH
Sbjct: 363 SVLIFRNPVTVMGITGFAVTIMGVLLYSEAKKRSKVTSH 479
>TC19748 weakly similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpyruvate
translocator protein-like, partial (22%)
Length = 427
Score = 169 bits (428), Expect = 6e-43
Identities = 87/125 (69%), Positives = 98/125 (77%), Gaps = 1/125 (0%)
Frame = +3
Query: 1 MVEAQTWTTRRMSNPRLHTLDTNDQLQLDIPQTPPSDQRN-NGSNINNNNLVTTSLIIAS 59
MVEAQTWTTRRMSNPRL T D++ L+IP TPP D R+ + + + T+ IIAS
Sbjct: 63 MVEAQTWTTRRMSNPRL----TADEVALEIPATPPGDVRHWQPFTSHFSPAMVTAAIIAS 230
Query: 60 WYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIHSKKQF 119
WY SNIGVLLLNKYLLSFYGYRYPIFLTMLHM+SC+AYSY AIN + VP Q IHSKKQF
Sbjct: 231 WYLSNIGVLLLNKYLLSFYGYRYPIFLTMLHMISCSAYSYGAINFLDLVPLQHIHSKKQF 410
Query: 120 LKIFA 124
LKIFA
Sbjct: 411 LKIFA 425
>AW720113
Length = 512
Score = 135 bits (340), Expect = 1e-32
Identities = 65/113 (57%), Positives = 82/113 (72%)
Frame = +2
Query: 39 RNNGSNINNNNLVTTSLIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYS 98
R N + + + T ++++WY SNIGVLLLNKYLLS YG++YPIFLTM HM +C+ +S
Sbjct: 173 RRNSTKMKGKTRLFTISLVSAWYSSNIGVLLLNKYLLSNYGFKYPIFLTMCHMTACSLFS 352
Query: 99 YAAINVVQFVPYQQIHSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAI 151
Y AI ++ VP Q I S+ QF KI LS +FC SVV GN SLRYLPVSFNQA+
Sbjct: 353 YVAIAWLKVVPLQTIRSRVQFFKIATLSLVFCVSVVFGNISLRYLPVSFNQAV 511
>TC15637 similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpyruvate
translocator protein-like, partial (32%)
Length = 680
Score = 135 bits (339), Expect = 1e-32
Identities = 69/103 (66%), Positives = 84/103 (80%), Gaps = 1/103 (0%)
Frame = +2
Query: 253 IEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTKHTSALTLQVLGNAKAAVA 312
+E NV IT+ AR D I++ L+ N+T+AY VNLTNFLVTKHTSALTLQVLGNAK AVA
Sbjct: 2 MEENVVGITLALARDDVKIIWYLLFNSTLAYFVNLTNFLVTKHTSALTLQVLGNAKGAVA 181
Query: 313 AVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAKKRSK-GASH 354
VVS+LIF+NPV+V GM G+ +T+ GV+LYSEAKKRSK G+ H
Sbjct: 182 VVVSILIFKNPVSVTGMMGYALTVFGVILYSEAKKRSK*GSDH 310
>BP046819
Length = 459
Score = 107 bits (268), Expect = 2e-24
Identities = 53/92 (57%), Positives = 67/92 (72%)
Frame = -3
Query: 257 VFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVVS 316
V +T+ A+ + LL+ N+ AY NLT FLVTKHTSALTLQVLGNAK AVA V+S
Sbjct: 454 VVDVTLTLAKDHKSMWILLLLNSVTAYAANLTXFLVTKHTSALTLQVLGNAKGAVAVVIS 275
Query: 317 VLIFRNPVTVMGMTGFGITIMGVVLYSEAKKR 348
+LIFRNPVT +GM G+ +T++GV Y E K+R
Sbjct: 274 ILIFRNPVTFVGMAGYTVTVLGVAAYGETKRR 179
>TC14096 homologue to UP|CPTR_PEA (P21727) Triose phosphate/phosphate
translocator, chloroplast precursor (CTPT) (P36) (E30),
partial (96%)
Length = 1564
Score = 92.0 bits (227), Expect = 1e-19
Identities = 78/304 (25%), Positives = 133/304 (43%), Gaps = 6/304 (1%)
Frame = +1
Query: 51 VTTSLIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPY 110
+ T +WYF N+ +LNK + +++ Y P F++++H+ Y + V +P
Sbjct: 340 LVTGFFFFTWYFLNVIFNILNKKIYNYFPY--PYFVSVIHLFVGVVYCLVSWTVG--LPK 507
Query: 111 QQIHSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKK 170
+ Q + ++ V N S + VSF + A PFF A + I +
Sbjct: 508 RAPIDSNQLKLLIPVAVCHALGHVTSNVSFAAVAVSFTHTVKALEPFFNAAASQFILGQS 687
Query: 171 ETAEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLH 230
++L+L PVV+G+ +++ +E F+ GF+ + S +S+ +T +
Sbjct: 688 IPITLWLSLAPVVIGVSMASLTELSFNWLGFISAMISNISFTYRSIYSKKAMTD----MD 855
Query: 231 SMNLLLYMAPLAAMILLPVTLYIEGNV-----FAITIEKARSDPFIVFLL-IGNATVAYL 284
S NL Y++ +A ++ LP + +EG V F I K F+ L +G Y
Sbjct: 856 STNLYAYISIIALIVCLPPAIILEGPVLLKHGFNDAIAKVGLVKFVTDLFWVGMFYHLYN 1035
Query: 285 VNLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSE 344
TN L + + LT V K S++IF N ++ G I I GV +YS
Sbjct: 1036QVATNTL--ERVAPLTHAVGNVLKRVFVIGFSIIIFGNRISTQTGIGTAIAIAGVAIYSL 1209
Query: 345 AKKR 348
K R
Sbjct: 1210IKAR 1221
>BP049869
Length = 532
Score = 73.2 bits (178), Expect = 6e-14
Identities = 49/178 (27%), Positives = 91/178 (50%), Gaps = 4/178 (2%)
Frame = -2
Query: 62 FSNIGVLLLNKYLLS--FYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIHSKKQF 119
F + V++ NKY+L Y + YPI LTM+HM C++ +Y + V + V + +
Sbjct: 531 FLSFTVIVYNKYILDRKMYNWPYPISLTMIHMGFCSSLAYLLVRVFKLVEPVSMSRELYL 352
Query: 120 LKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKET--AEVYL 177
+ + A++ S+ N++ YL VSF Q + A P A+++ + KKE+ E
Sbjct: 351 KSVVPIGALYSLSLWFSNSAYIYLSVSFIQMLKALMP--VAVYSIGVLFKKESFRNETMA 178
Query: 178 ALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNLL 235
++ + +G+ V+ E F +FG + + + A A + V+ I+L S+ L+ + L
Sbjct: 177 NMVSISMGVAVAAYGEAKFDVFGVSLQLLAVAFEATRLVLIQILLYSKGISLNPITSL 4
>TC11969 similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpyruvate
translocator protein-like, partial (14%)
Length = 503
Score = 68.2 bits (165), Expect = 2e-12
Identities = 32/43 (74%), Positives = 38/43 (87%)
Frame = +1
Query: 308 KAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAKKRSK 350
K AVA VVS+LIFRNPV+V GM G+ +T++GVVLYSEAKKRSK
Sbjct: 1 KGAVAVVVSILIFRNPVSVTGMMGYSLTVLGVVLYSEAKKRSK 129
>TC13454 homologue to UP|O64910 (O64910)
Glucose-6-phosphate/phosphate-translocator precursor,
partial (56%)
Length = 674
Score = 62.4 bits (150), Expect = 1e-10
Identities = 47/205 (22%), Positives = 94/205 (44%), Gaps = 3/205 (1%)
Frame = +2
Query: 122 IFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAEVYLALLP 181
+F ++ V S+ + VSF I + P F+ + + I + A+VYL+LLP
Sbjct: 65 LFPVAVAHTIGHVAATVSMSKVAVSFTHIIKSGEPAFSVLVSRFILGESFPAQVYLSLLP 244
Query: 182 VVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNLLLYMAPL 241
++ G ++ +E F++ GF+ + S +++ + + + MN ++ L
Sbjct: 245 IIGGCALAAVTELNFNMVGFMGAMISNLAFVFRNIFSK--KGMKGKSVSGMNYYACLSIL 418
Query: 242 AAMILLPVTLYIEG-NVFAITIEKARSD--PFIVFLLIGNATVAYLVNLTNFLVTKHTSA 298
+ IL P + +EG ++A + A S P +V+ + + +L N +++ S
Sbjct: 419 SLAILTPFAIAVEGPQMWAAGWQTAVSQVGPQLVWWVAAQSVFYHLYNQVSYMSLDEISP 598
Query: 299 LTLQVLGNAKAAVAAVVSVLIFRNP 323
LT + K V S++IF+ P
Sbjct: 599 LTFSIGNTMKRISVIVSSIIIFQTP 673
>TC15915 similar to UP|Q9SFE9 (Q9SFE9) T26F17.9 (GONST5 golgi nucleotide
sugar transporter), partial (38%)
Length = 545
Score = 61.2 bits (147), Expect = 2e-10
Identities = 34/93 (36%), Positives = 53/93 (56%), Gaps = 3/93 (3%)
Frame = +2
Query: 253 IEGNVFAITIEKARSDPF---IVFLLIGNATVAYLVNLTNFLVTKHTSALTLQVLGNAKA 309
+EGN IE + P+ + ++ + +A+ +N + F V T+A+T V GN K
Sbjct: 2 LEGNG---VIEWLNTHPYPWLALTIIFSSGVLAFCLNFSIFYVIHSTTAVTFNVAGNLKV 172
Query: 310 AVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLY 342
AVA +VS LIFRNP++ + G GIT++G Y
Sbjct: 173 AVAVLVSWLIFRNPISYLNAVGCGITLVGCTFY 271
>TC17295 similar to UP|Q94JT2 (Q94JT2) AT5g17630/K10A8_110, partial (28%)
Length = 566
Score = 54.7 bits (130), Expect = 2e-08
Identities = 33/114 (28%), Positives = 57/114 (49%), Gaps = 5/114 (4%)
Frame = +3
Query: 237 YMAPLAAMILLPVTLYIEGNVFAITIEKA-----RSDPFIVFLLIGNATVAYLVNLTNFL 291
++ L+ L PV +++EG+ + KA + F +++L+ + +L N +++
Sbjct: 9 FITILSLFYLFPVAIFVEGSQWIPGYYKAIETIGKPSTFYIWVLV-SGVFYHLYNQSSYQ 185
Query: 292 VTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEA 345
S LT V K V V ++L+FRNPV + G I I+G LYS+A
Sbjct: 186 ALDEISPLTFSVGNTMKRVVVIVATILVFRNPVRPLNGLGSAIAILGTFLYSQA 347
>TC10652 similar to UP|Q9SFE9 (Q9SFE9) T26F17.9 (GONST5 golgi nucleotide
sugar transporter), partial (28%)
Length = 595
Score = 54.3 bits (129), Expect = 3e-08
Identities = 27/62 (43%), Positives = 37/62 (59%)
Frame = +3
Query: 281 VAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVV 340
+A+ +N + F V A+T V GN K AVA +VS LIFRNP++ M G IT++G
Sbjct: 9 LAFCLNFSIFYVIHSPPAVTFNVAGNLKVAVAVLVSWLIFRNPISYMNSIGCAITLIGCT 188
Query: 341 LY 342
Y
Sbjct: 189 FY 194
>TC12775 similar to UP|Q9FG70 (Q9FG70) Gb|AAF04433.1, partial (45%)
Length = 712
Score = 46.2 bits (108), Expect = 8e-06
Identities = 40/184 (21%), Positives = 84/184 (44%), Gaps = 1/184 (0%)
Frame = +2
Query: 161 IFAFLITCKKETAEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGI 220
+ F++ K + + LAL V G+ V+T ++ F+ FG +V V A+ ++
Sbjct: 8 VAXFILFGKTISFKKVLALAVVSAGVAVATVTDLXFNFFGAIVAVIWIIPSAINXILWST 187
Query: 221 ILTSEAEKLHSMNLLLYMAPLAAMILLPVTLYIEG-NVFAITIEKARSDPFIVFLLIGNA 279
+ + ++ L+ P+ L+ + +I+ + + + S +V L+G
Sbjct: 188 L--QQQGNWTALALMWKTTPITVFFLVALMPWIDPPGILSFKWDVNNSTTIMVSALLG-- 355
Query: 280 TVAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGV 339
+L+ + L TSA T VLG K V + L+F++ V+ ++G + + G+
Sbjct: 356 ---FLLQWSGALALGATSATTHVVLGQFKTCVILLGGYLLFKSDPGVISISGAVVALSGM 526
Query: 340 VLYS 343
+Y+
Sbjct: 527 SIYT 538
>TC14931 similar to UP|Q9M9Z3 (Q9M9Z3) F4H5.5 protein, partial (55%)
Length = 820
Score = 45.8 bits (107), Expect = 1e-05
Identities = 25/104 (24%), Positives = 51/104 (49%)
Frame = +1
Query: 240 PLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTKHTSAL 299
P A LL Y++ + + + + + ++ + ++ VN + FLV TS +
Sbjct: 10 PYQAATLLICGPYLDKLLTNLNVFAFKYTTQVTAFIVLSCLISISVNFSTFLVIGKTSPV 189
Query: 300 TLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYS 343
T QVLG+ K + ++ R+P + + G + ++G++LYS
Sbjct: 190 TYQVLGHLKTCLVLAFGYILLRDPFSWRNILGILVAMIGMILYS 321
>TC12333 similar to GB|AAP42755.1|30984584|BT008742 At2g30460 {Arabidopsis
thaliana;}, partial (20%)
Length = 474
Score = 37.0 bits (84), Expect = 0.005
Identities = 23/76 (30%), Positives = 36/76 (47%), Gaps = 5/76 (6%)
Frame = +2
Query: 281 VAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVV 340
+A VN + FLV TS +T VLG+ + + + +P T + G I + G+
Sbjct: 2 IAVSVNFSTFLVIGKTSPVTYPVLGHLQTCLVLGFGYTLLHDPFTGRNILGILIAVSGMG 181
Query: 341 LYS-----EAKKRSKG 351
LYS E +K+ G
Sbjct: 182 LYSYFCTQENEKKQSG 229
>BP071016
Length = 398
Score = 30.4 bits (67), Expect = 0.47
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 1/40 (2%)
Frame = +1
Query: 79 GYRYPIFLTMLHMLSCAAY-SYAAINVVQFVPYQQIHSKK 117
GYRYP ++ M H SCA+Y S + + Q V + S K
Sbjct: 58 GYRYPQYVHMYHYRSCASYISNT*VPIFQLVTTRHKRSNK 177
>TC8964 weakly similar to UP|Q94F54 (Q94F54) AT3g15070/K15M2_22, partial
(19%)
Length = 594
Score = 28.5 bits (62), Expect = 1.8
Identities = 13/47 (27%), Positives = 23/47 (48%)
Frame = +2
Query: 84 IFLTMLHMLSCAAYSYAAINVVQFVPYQQIHSKKQFLKIFALSAIFC 130
+FL +H+L C Y Y IN + P + + + + A+ +FC
Sbjct: 419 LFLNRIHILFCNLYMYLHINRQSYCPTPLLLAPEPCIYQQAVQTLFC 559
>TC7831 similar to UP|Q8H946 (Q8H946) Phosphoenolpyruvate carboxylase ,
partial (6%)
Length = 489
Score = 27.3 bits (59), Expect = 4.0
Identities = 15/66 (22%), Positives = 32/66 (47%), Gaps = 1/66 (1%)
Frame = +1
Query: 64 NIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQF-VPYQQIHSKKQFLKI 122
NI L ++ L+ F+ +P+F+ + C YAA+ Q +P+Q +H + +
Sbjct: 226 NIVASLPSECLVFFFSQSHPLFVITMFSPLCCMIRYAALEFHQRPLPHQ*VHPNNSSVSL 405
Query: 123 FALSAI 128
+ ++
Sbjct: 406 RKICSV 423
>BI418051
Length = 668
Score = 26.9 bits (58), Expect = 5.2
Identities = 20/52 (38%), Positives = 30/52 (57%)
Frame = +3
Query: 140 LRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAEVYLALLPVVLGIVVSTN 191
L YLP+S + AT F+A+F+F + +K T L L VVL + +ST+
Sbjct: 447 LLYLPLSTYVLLCATQLAFSALFSFFLNSQKFTK---LILNSVVL-LTISTS 590
>BP036253
Length = 501
Score = 26.6 bits (57), Expect = 6.8
Identities = 12/37 (32%), Positives = 18/37 (48%)
Frame = +2
Query: 190 TNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEA 226
TN +P HL F + + ++ LKS + SEA
Sbjct: 44 TNMKPNIHLISFFILISQSSKTCLKSTILNSRTISEA 154
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.325 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,592,543
Number of Sequences: 28460
Number of extensions: 76699
Number of successful extensions: 606
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 598
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 602
length of query: 354
length of database: 4,897,600
effective HSP length: 91
effective length of query: 263
effective length of database: 2,307,740
effective search space: 606935620
effective search space used: 606935620
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC137510.9


